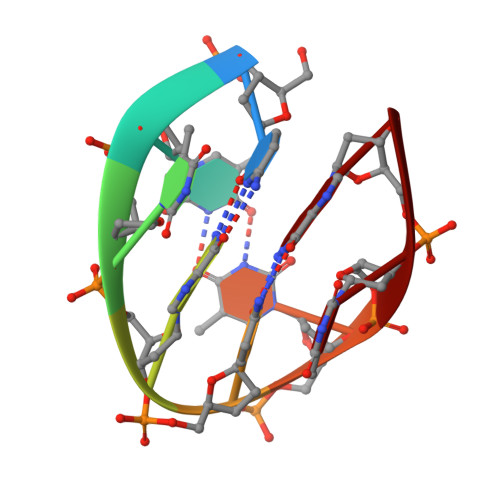
Unprecedented hydrophobic stabilizations from a reverse wobble T·T mispair in DNA minidumbbell.
Guo, P., Lam, S.L.(2020) J Biomol Struct Dyn 38: 1946-1953
- PubMed: 31107180
- DOI: https://doi.org/10.1080/07391102.2019.1621211
- Primary Citation of Related Structures:
6J37 - PubMed Abstract:
Minidumbbell (MDB) is a newly found non-B DNA structure formed by short single-strand sequences. Up to now, three MDBs have been reported to form at neutral pH by sequences containing two repeats of TTTA, CCTG and CTTG. Among them, the thermodynamically less stable TTTA and CCTG MDBs have been proposed to be the structural intermediates that cause TTTA and CCTG repeat expansions during DNA replication in Staphylococcus aureus pathogen and myotonic dystrophy type 2 patients, respectively. Although the CTTG MDB has a melting temperature of at least 13 °C higher than those of the other two, no CTTG repeat expansion has ever been reported in any genomes. In this study, we successfully determined the solution structure of the CTTG MDB and observed for the first time the formation of a reverse wobble T·T mispair with two symmetric hydrogen bonds. More importantly, we identified unprecedented hydrophobic interactions between the two methyl groups of this T·T mispair and the four 2'-methylene groups of their nearby loop-closing base pair residues. These stabilizations account for the substantial increase in the MDB thermodynamic stability which may govern the occurrence of repeat expansions.Communicated by Ramaswamy H. Sarma [Formula: see text].
Organizational Affiliation:
Department of Chemistry, The Chinese University of Hong Kong, Shatin, New Territories, Hong Kong.