Structural determination of group A Streptococcal surface dehydrogenase and characterization of its interaction with urokinase-type plasminogen activator receptor.
Li, R., Liang, C., Jiang, L., Yuan, C., Huang, M.(2019) Biochem Biophys Res Commun 510: 539-544
- PubMed: 30737033
- DOI: https://doi.org/10.1016/j.bbrc.2019.01.102
- Primary Citation of Related Structures:
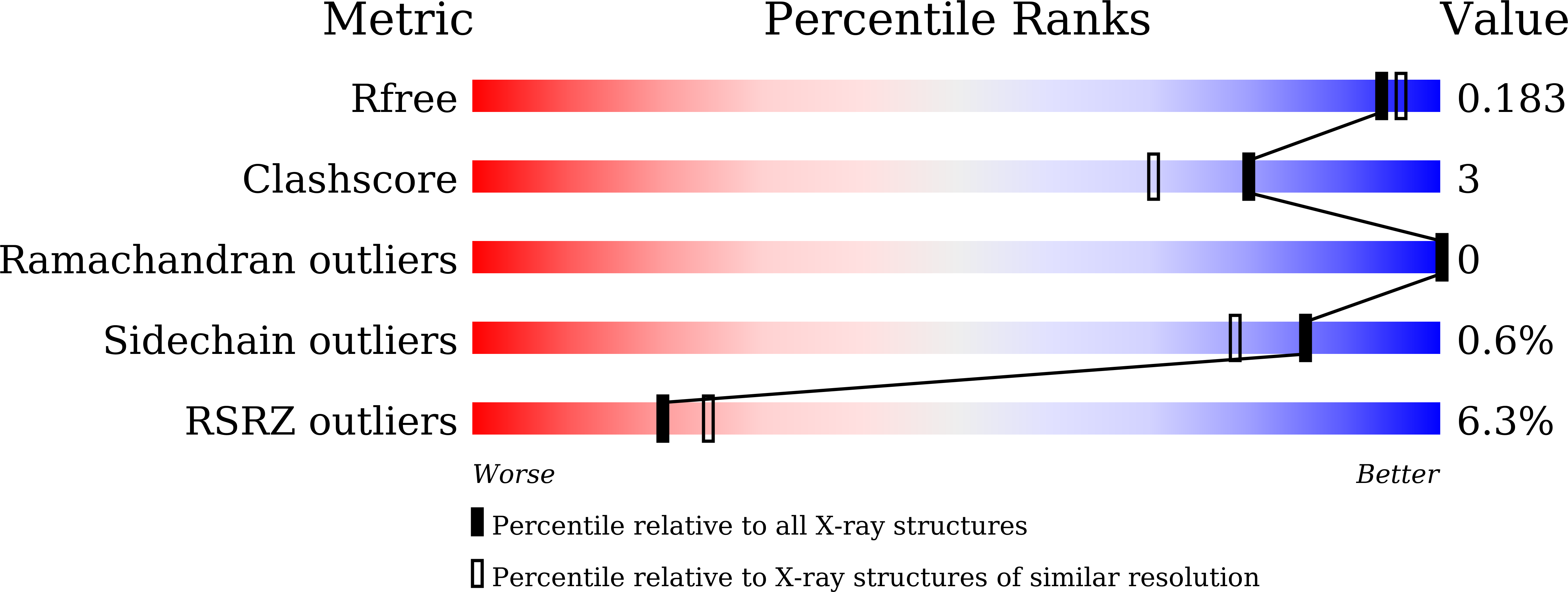
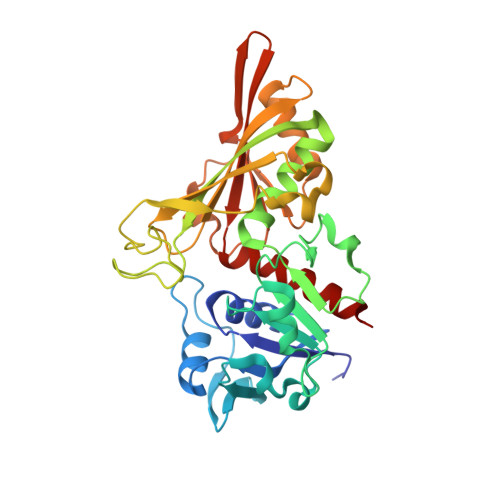
6ITE - PubMed Abstract:
Streptococcus pyogenes (group A Streptococcus, GAS) has caused a wide variety of human diseases. Its multifunctional surface dehydrogenase (SDH) is crucial for GAS life cycle. Furthermore, GAS infection into human pharyngeal cells has been previously shown to be mediated by the interaction between SDH and host urokinase-type plasminogen activator receptor (uPAR). However, the structural information of SDH remains to be elucidated and there are few detailed studies to characterize its interaction with uPAR. In-depth research on these issues will provide potential targets and strategies for combating GAS. Here, we prepared recombinant SDH tetramer in Escherichia coli BL21 (DE3) cells. After purification and crystallization, we determined its crystal structure at 1.74 Å. The unique characteristics might be potentially explored as drug targets or vaccine immunogen. We subsequently performed gel filtration chromatography, native-polyacrylamide gel electrophoresis (PAGE) and in vitro pull-down analyses. The results showed that their interaction was too weak to form stable complexes and the role of uPAR involved in GAS infection needs further demonstration. Altogether the current work provides the first view of SDH and deepens the knowledge of GAS infection.
Organizational Affiliation:
Key Laboratory of Animal Immunology of the Ministry of Agriculture, Henan Provincial Key Laboratory of Animal Immunology, Henan Academy of Agricultural Sciences, Zhengzhou, 450002, Henan, China. Electronic address: lirui860620@sina.com.