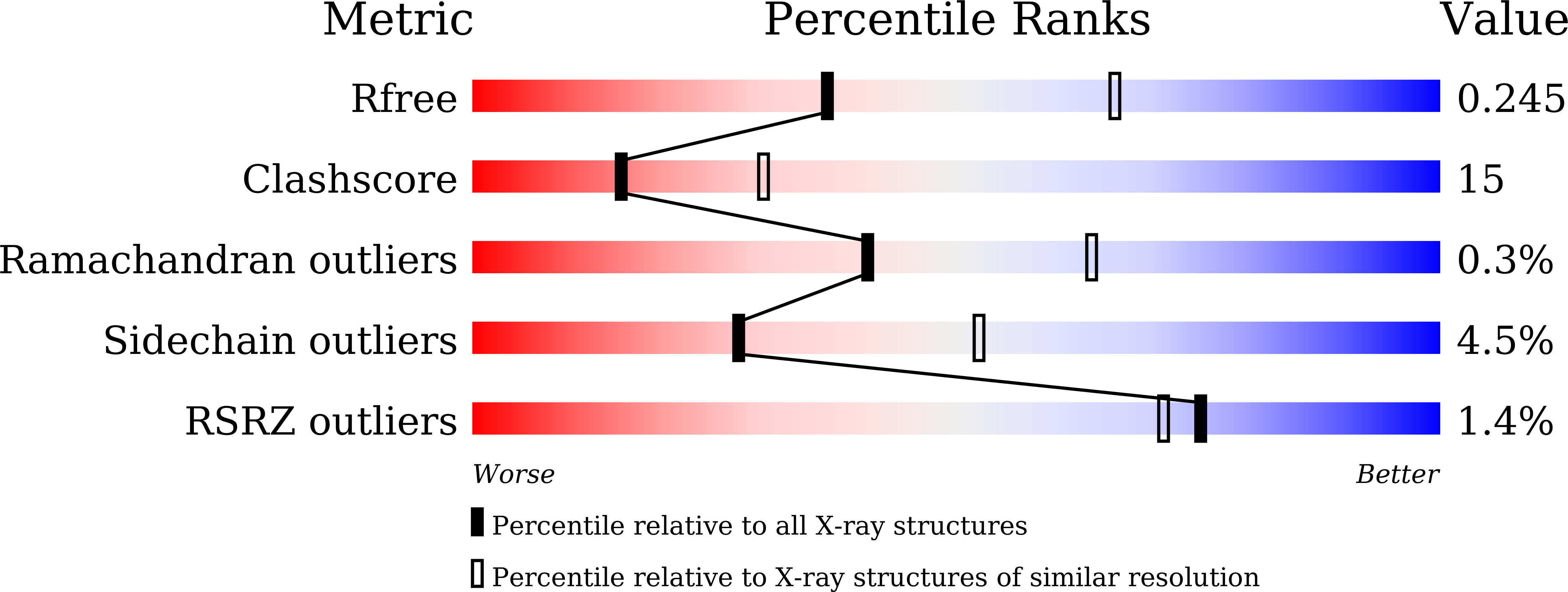
Crystal structure of GAPDH of Streptococcus agalactiae and characterization of its interaction with extracellular matrix molecules
Nagarajan, R., Sankar, S., Ponnuraj, K.(2018) Microb Pathog 127: 359-367
- PubMed: 30553015
- DOI: https://doi.org/10.1016/j.micpath.2018.12.020
- Primary Citation of Related Structures:
6IEP - PubMed Abstract:
GAPDH being a key enzyme in the glycolytic pathway is one of the surface adhesins of many Gram-positive bacteria including Streptococcus agalactiae. This anchorless adhesin is known to bind to host plasminogen (PLG) and fibrinogen (Fg), which enhances the virulence and modulates the host immune system. The crystal structure of the recombinant GAPDH from S. agalactiae (SagGAPDH) was determined at 2.6 Å resolution by molecular replacement. The structure was found to be highly conserved with a typical NAD binding domain and a catalytic domain. In this paper, using biolayer interferometry studies, we report that the multifunctional SagGAPDH enzyme binds to a variety of host molecules such as PLG, Fg, laminin, transferrin and mucin with a K D value of 4.4 × 10 -7 M, 9.8 × 10 -7 M, 1 × 10 -5 M, 9.7 × 10 -12 M and 1.4 × 10 -7 M respectively. The ligand affinity blots reveal that SagGAPDH binds specifically to α and β subunits of Fg and the competitive binding ELISA assay reveals that the Fg and PLG binding sites on GAPDH does not overlap each other. The PLG binding motif of GAPDH varies with organisms, however positively charged residues in the hydrophobic surroundings is essential for PLG binding. The lysine analogue competitive binding assay and lysine succinylation experiments deciphered the role of SagGAPDH lysines in PLG binding. On structural comparison with S. pneumoniae GAPDH, K171 of SagGAPDH is being predicted to be involved in PLG binding. Further SagGAPDH exhibited enzymatic activity in the presence of Fg, PLG and transferrin. This suggests that these host molecules does not mask the active site and bind at some other region of GAPDH.
Organizational Affiliation:
Centre of Advanced Study in Crystallography and Biophysics, University of Madras, Guindy Campus, Chennai, 600025, India.