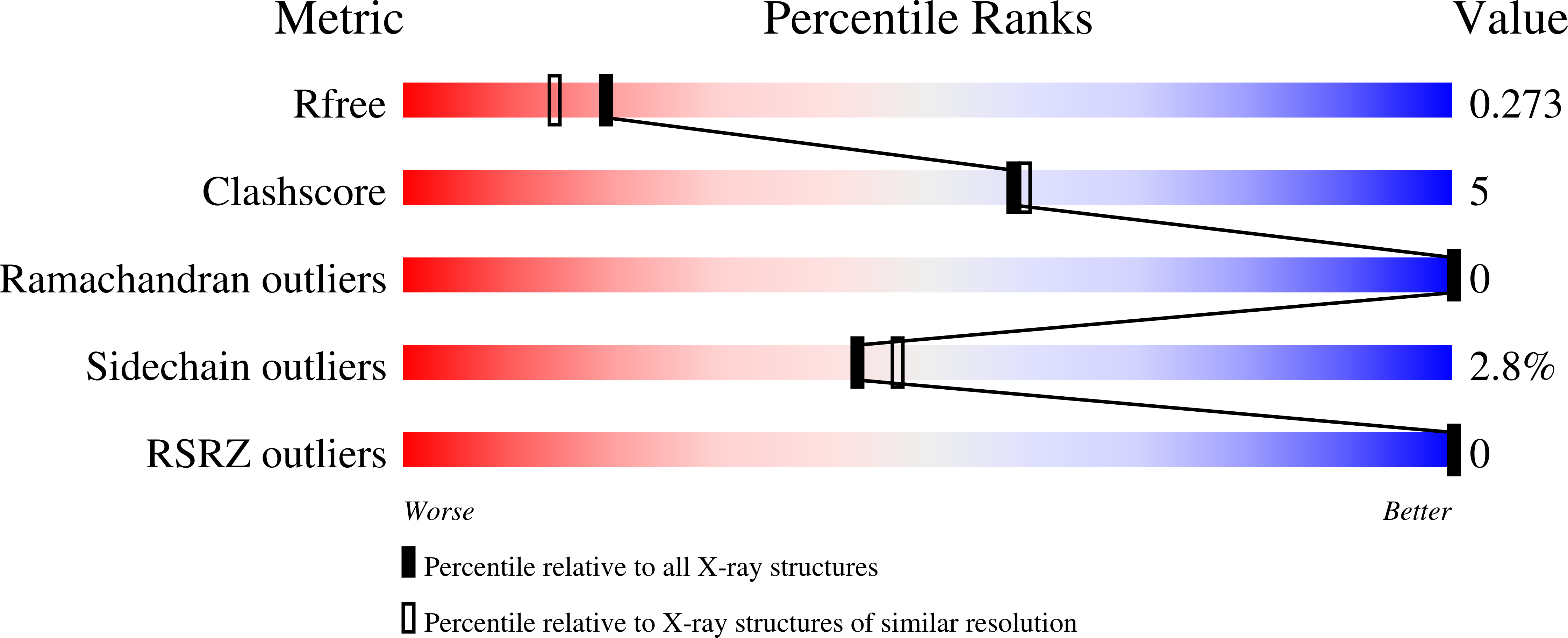
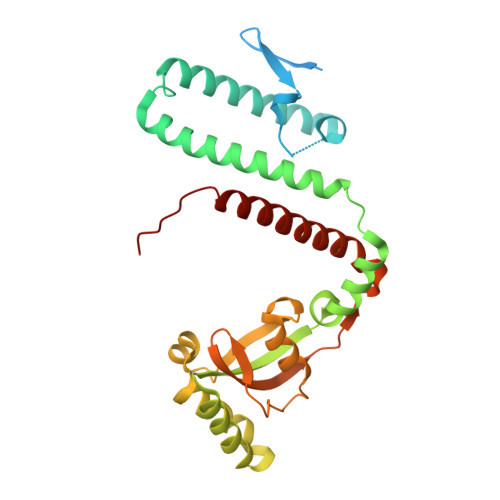
Structural flexibility in the Helicobacter pylori peptidyl-prolyl cis,trans-isomerase HP0175 is achieved through an extension of the chaperone helices.
Yaseen, A., Audette, G.F.(2018) J Struct Biol 204: 261-269
- PubMed: 30179659
- DOI: https://doi.org/10.1016/j.jsb.2018.08.017
- Primary Citation of Related Structures:
6BHF - PubMed Abstract:
Helicobacter pylori infects the gastric epithelium of half the global population, where infections can persist into adenocarcinomas and peptic ulcers. H. pylori secretes several proteins that lend to its pathogenesis and survival including VacA, CagA, γ-glutamyltransferase and HP0175. HP0175, also known as HpCBF2, classified as a peptidyl-prolyl cis,trans-isomerase, has been shown to induce apoptosis through a cascade of mechanisms initiated though its interaction with toll like receptor 4 (TLR4). Here, we report the structure of apo-HP0175 at 2.09 Å with a single monomer in the asymmetric unit. Chromatographic, light scattering and mass spectrometric analysis of HP0175 in solution indicate that the protein is mainly monomeric under low salt conditions, while increasing ionic interactions facilitates protein dimerization. A comparison of the apo-HP0175 structure to that of the indole-2-carboxylic acid-bound form shows movement of the N- and C-terminal helices upon interaction of the catalytic residues in the binding pocket. Helix extension of the N/C chaperone domains between apo and I2CA-bound HP0175 supports previous findings in parvulin PPIases for their role in protein stabilization (and accommodation of variable protein lengths) of those undergoing catalysis.
Organizational Affiliation:
Department of Chemistry, York University, Toronto M3J 1P3, Canada.