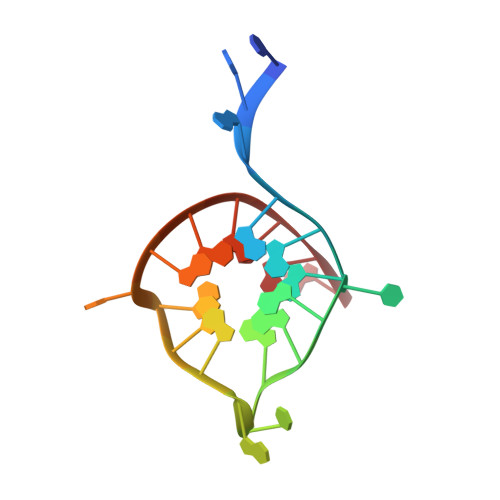
Crystal structure of the major quadruplex formed in the promoter region of the human c-MYC oncogene.
Stump, S., Mou, T.C., Sprang, S.R., Natale, N.R., Beall, H.D.(2018) PLoS One 13: e0205584-e0205584
- PubMed: 30312328
- DOI: https://doi.org/10.1371/journal.pone.0205584
- Primary Citation of Related Structures:
6AU4 - PubMed Abstract:
The c-MYC oncogene mediates multiple tumor cell survival pathways and is dysregulated or overexpressed in the majority of human cancers. The NHE III1 region of the c-MYC promoter forms a DNA quadruplex. Stabilization of this structure with small molecules has been shown to reduce expression of c-MYC, and targeting the c-MYC quadruplex has become an emerging strategy for development of antitumor compounds. Previous solution NMR studies of the c-MYC quadruplex have assigned the major conformer and topology of this important target, however, regions outside the G-quartet core were not as well-defined. Here, we report a high-resolution crystal structure (2.35 Å) of the major quadruplex formed in the NHE III1 region of the c-MYC promoter. The crystal structure is in general agreement with the solution NMR structure, however, key differences are observed in the position of nucleotides outside the G-quartet core. The crystal structure provides an alternative model that, along with comparisons to other reported quadruplex crystal structures, will be important to the rational design of selective compounds. This work will aid in development of ligands to target the c-MYC promoter quadruplex with the goal of creating novel anticancer therapies.
Organizational Affiliation:
Center for Environmental Health Sciences, Department of Biomedical and Pharmaceutical Sciences, University of Montana, Missoula, Montana, United States of America.