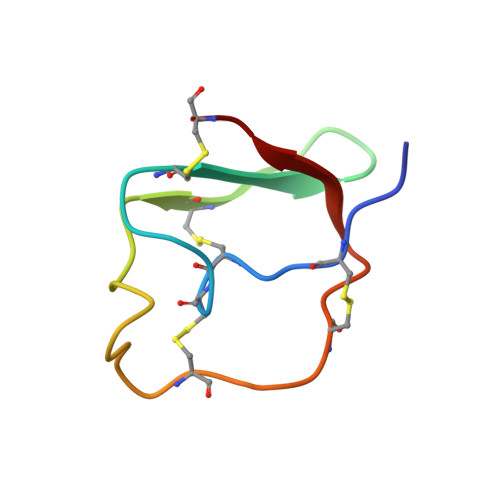
NMR structure and dynamics of inhibitory repeat domain variant 12, a plant protease inhibitor from Capsicum annuum, and its structural relationship to other plant protease inhibitors.
Gartia, J., Anangi, R., Joshi, R.S., Giri, A.P., King, G.F., Barnwal, R.P., Chary, K.V.R.(2019) J Biomol Struct Dyn : 1-10
- PubMed: 31038412
- DOI: https://doi.org/10.1080/07391102.2019.1607559
- Primary Citation of Related Structures:
5ZFO - PubMed Abstract:
Although several plant protease inhibitors have been structurally characterized using X-ray crystallography, very few have been studied using NMR techniques. Here, we report an NMR study of the solution structure and dynamics of an inhibitory repeat domain (IRD) variant 12 from the wound-inducible Pin-II type proteinase inhibitor from Capsicum annuum . IRD variant 12 (IRD12) showed strong anti-metabolic activity against the Lepidopteran insect pest, Helicoverpa armigera . The NMR-derived three-dimensional structure of IRD12 reveals a three-stranded anti-parallel β-sheet rigidly held together by four disulfide bridges and shows structural homology with known IRDs. It is interesting to note that the IRD12 structure containing ∼75% unstructured part still shows substantial amount of rigidity of N-H bond vectors with respect to its molecular motion.Communicated by Ramaswamy H. Sarma.
Organizational Affiliation:
Center for Interdisciplinary Sciences, Tata Institute of Fundamental Research, Gopanpally, Hyderabad, India.