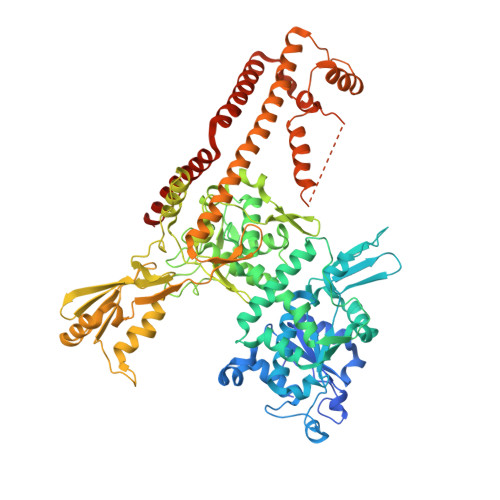
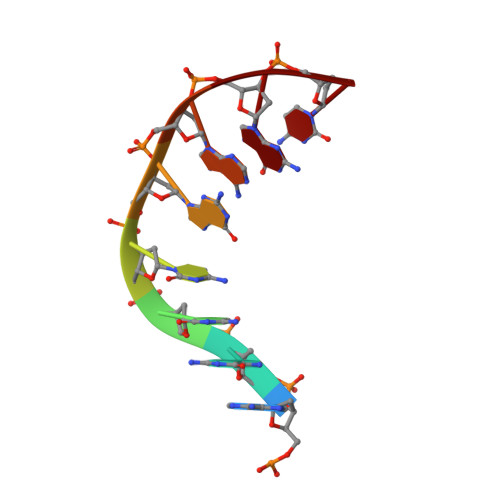
Increasing the distance between two monomers of topoisomerase II beta under the action of antitumor agent 4 beta-sulfur-(benzimidazole) 4'-demethylepipodophyllotoxin.
Sun, L.Y., Zhu, L.W., Tang, Y.J.(2018) Sci Rep 8: 14949-14949
- PubMed: 30297860
- DOI: https://doi.org/10.1038/s41598-018-33366-2
- Primary Citation of Related Structures:
5ZAD - PubMed Abstract:
Topoisomerases II (Top2s) are a group of essential enzymes involved in replication, transcription, chromosome condensation, and segregation via altering DNA topology. The mechanism of the Top2s poisons such as etoposide (VP-16) was reported as stabilizing the Top2-DNA complex and engendering permanent DNA breakage. As the structurally similar compound of VP-16, a novel 4β-sulfur-substituted 4'-demethylepipodophyllotoxin (DMEP) derivative (compound C-Bi) with superior antitumor activity was developed in our previous study. To understand the structural basis of the compound action, the crystal structure (2.54 Å) of human Top2 β-isoform (hTop2β) cleavage complexes stabilized by compound C-Bi was determined. However, compound C-Bi was not visible in the crystal structure. Through the comparison of the structures of hTop2β-DNA-etoposide ternary complex and hTop2β-DNA binary complex, it could be observed that the distance between drug-binding sites Arg503 of the two monomers was 26.62 Å in hTop2β-DNA-etoposide ternary complex and 34.54 Å in hTop2β-DNA binary complex, respectively. Significant twist were observed in the DNA chains of binary complex. It suggested that compound C-Bi played antitumor roles through increasing spacing of hTop2β monomers. The changes in hTop2β structure further caused double changes in the torsional direction and migration distance of the DNA chains, resulting in impeding religation of DNA.
Organizational Affiliation:
Hubei Key Laboratory of Industrial Microbiology, Hubei Provincial Cooperative Innovation Center of Industrial Fermentation, Key Laboratory of Fermentation Engineering (Ministry of Education), Hubei University of Technology, Wuhan, 430068, China.