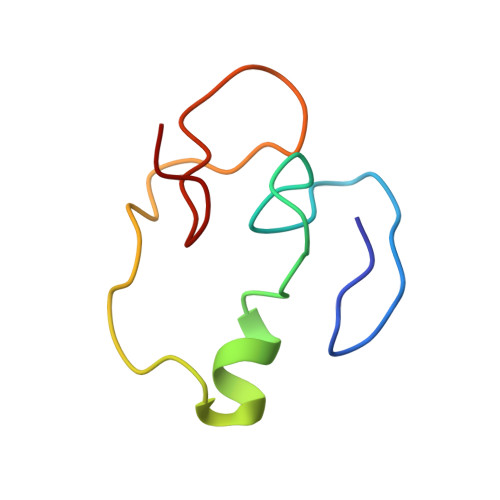
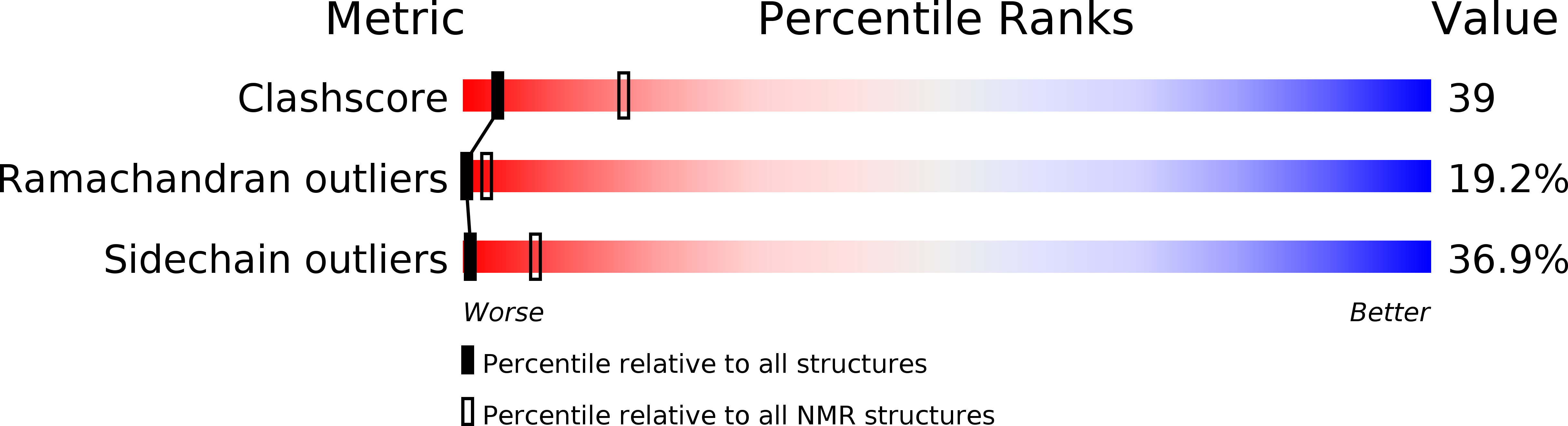Solution structure of the PHD finger from the human KIAA1045 protein
Miyamoto, K., Yamashita, A., Saito, K.(2018) Protein Sci 27: 987-992
- PubMed: 29430827
- DOI: https://doi.org/10.1002/pro.3389
- Primary Citation of Related Structures:
5XHT - PubMed Abstract:
Cross-brace structural motifs are required as a scaffold to design artificial RING fingers (ARFs) that function as ubiquitin ligase (E3) in ubiquitination and have specific ubiquitin-conjugating enzyme (E2)-binding capabilities. The Simple Modular Architecture Research Tool database predicted the amino acid sequence 131-190 (KIAA1045ZF) of the human KIAA1045 protein as an unidentified structural region. Herein, the stoichiometry of zinc ions estimated spectrophotometrically by the metallochromic indicator revealed that the KIAA1045ZF motif binds to two zinc atoms. The structure of the KIAA1045ZF motif bound to the zinc atoms was elucidated at the atomic level by nuclear magnetic resonance. The actual structure of the KIAA1045ZF motif adopts a C 4 HC 3 -type PHD fold belonging to the cross-brace structural family. Therefore, the utilization of the KIAA1045ZF motif as a scaffold may lead to the creation of a novel ARF.
Organizational Affiliation:
Department of Pharmaceutical Health Care, Faculty of Pharmaceutical Sciences, Himeji Dokkyo University, Hyogo, Japan.