Improvement of enzyme activity of beta-1,3-1,4-glucanase from Paenibacillus sp. X4 by error-prone PCR and structural insights of mutated residues.
Baek, S.C., Ho, T.H., Lee, H.W., Jung, W.K., Gang, H.S., Kang, L.W., Kim, H.(2017) Appl Microbiol Biotechnol 101: 4073-4083
- PubMed: 28180917
- DOI: https://doi.org/10.1007/s00253-017-8145-4
- Primary Citation of Related Structures:
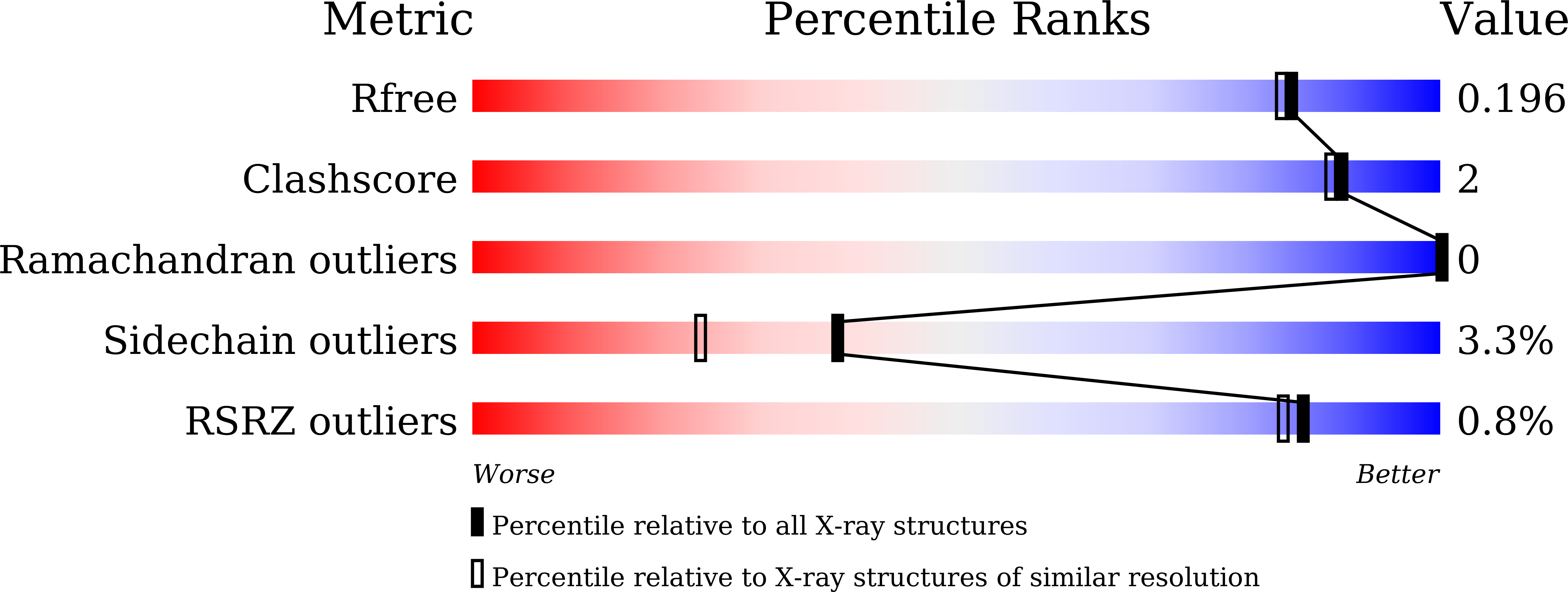
5XD0 - PubMed Abstract:
β-1,3-1,4-Glucanase (BGlc8H) from Paenibacillus sp. X4 was mutated by error-prone PCR or truncated using termination primers to improve its enzyme properties. The crystal structure of BGlc8H was determined at a resolution of 1.8 Å to study the possible roles of mutated residues and truncated regions of the enzyme. In mutation experiments, three clones of EP 2-6, 2-10, and 5-28 were finally selected that exhibited higher specific activities than the wild type when measured using their crude extracts. Enzyme variants of BG 2-6 , BG 2-10 , and BG 5-28 were mutated at two, two, and six amino acid residues, respectively. These enzymes were purified homogeneously by Hi-Trap Q and CHT-II chromatography. Specific activity of BG 5-28 was 2.11-fold higher than that of wild-type BG wt , whereas those of BG 2-6 and BG 2-10 were 0.93- and 1.19-fold that of the wild type, respectively. The optimum pH values and temperatures of the variants were nearly the same as those of BG wt (pH 5.0 and 40 °C, respectively). However, the half-life of the enzyme activity and catalytic efficiency (k cat /K m ) of BG 5-28 were 1.92- and 2.12-fold greater than those of BG wt at 40 °C, respectively. The catalytic efficiency of BG 5-28 increased to 3.09-fold that of BG wt at 60 °C. These increases in the thermostability and catalytic efficiency of BG 5-28 might be useful for the hydrolysis of β-glucans to produce fermentable sugars. Of the six mutated residues of BG 5-28 , five residues were present in mature BGlc8H protein, and two of them were located in the core scaffold of BGlc8H and the remaining three residues were in the substrate-binding pocket forming loop regions. In truncation experiments, three forms of C-terminal truncated BGlc8H were made, which comprised 360, 286, and 215 amino acid residues instead of the 409 residues of the wild type. No enzyme activity was observed for these truncated enzymes, suggesting the complete scaffold of the α 6 /α 6 -double-barrel structure is essential for enzyme activity.
Organizational Affiliation:
Department of Pharmacy, Sunchon National University, Suncheon, 57922, Republic of Korea.