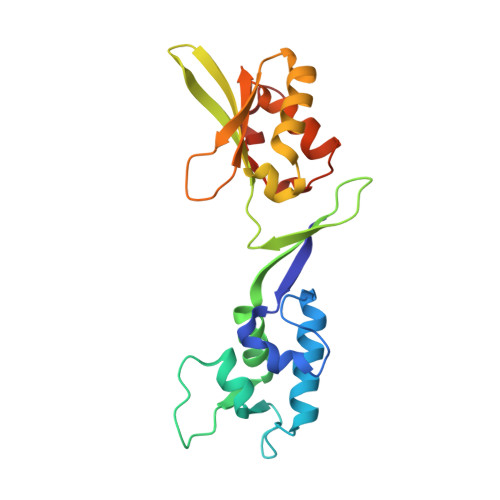
Crystal Structure of the C-terminal Domain of Human eIF2D and Its Implications on Eukaryotic Translation Initiation.
Vaidya, A.T., Lomakin, I.B., Joseph, N.N., Dmitriev, S.E., Steitz, T.A.(2017) J Mol Biol 429: 2765-2771
- PubMed: 28736176
- DOI: https://doi.org/10.1016/j.jmb.2017.07.015
- Primary Citation of Related Structures:
5W2F - PubMed Abstract:
Protein synthesis is a key process in all living organisms. In eukaryotes, initiation factor 2 (eIF2) plays an important role in translation initiation as it selects and delivers the initiator tRNA to the small ribosomal subunit. Under stress conditions, phosphorylation of the α-subunit of eIF2 downregulates cellular protein synthesis. However, translation of certain mRNAs continues via the eIF2D-dependent non-canonical initiation pathway. The molecular mechanism of this process remains elusive. In addition, eIF2D plays a role in translation re-initiation and ribosome recycling. Currently, there has been no structural information of eIF2D. We have now determined the crystal structure of the C-terminal domains of eIF2D at 1.4-Å resolution. One domain has the fold similar to that of eIF1, which is crucial for the scanning and initiation codon selection. The second domain has a known SWIB/MDM2 fold, which was not observed before in other translation initiation factors. Our structure reveals atomic details of inter-domain interactions in the C-terminal part of eIF2D and sheds light on the possible role of these domains in eIF2D during translation.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520-8114, USA; Howard Hughes Medical Institute, Yale University, New Haven, CT 06520-8114, USA.