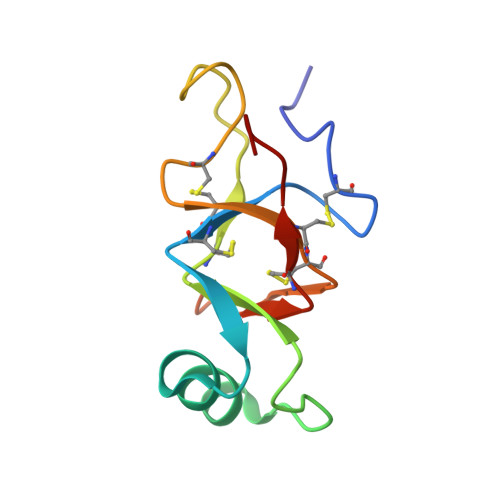
Characterization of the structure and self-assembly of two distinct class IB hydrophobins.
Vergunst, K.L., Kenward, C., Langelaan, D.N.(2022) Appl Microbiol Biotechnol 106: 7831-7843
- PubMed: 36329133
- DOI: https://doi.org/10.1007/s00253-022-12253-x
- Primary Citation of Related Structures:
5W0Y - PubMed Abstract:
Hydrophobins are small proteins secreted by fungi that accumulate at interfaces, modify surface hydrophobicity, and self-assemble into large amyloid-like structures. These unusual properties make hydrophobins an attractive target for commercial applications as emulsifiers and surface modifying agents. Hydrophobins have diverse sequences and tertiary structures, complicating attempts to characterize how they function. Here we describe the atomic resolution structure of the unusual hydrophobin SLH4 (86 aa, 8.4 kDa) and compare its function to another hydrophobin, SC16 (99 aa, 10.2 kDa). Despite containing only one charged residue, SLH4 has a similar structure to SC16 yet has strikingly different rodlet morphology, propensity to self-assemble, and preferred assembly conditions. Secondary structure analysis of both SC16 and SLH4 suggest that during rodlet formation residues in the first intercysteine loop undergo conformational changes. This work outlines a representative structure for class IB hydrophobins and illustrates how hydrophobin surface properties govern self-assembly, which provides context to rationally select hydrophobins for applications as surface modifiers. KEY POINTS: • The atomic-resolution structure of the hydrophobin SLH4 was determined using nuclear magnetic resonance spectroscopy. • The structure of SLH4 outlines a representative structure for class IB hydrophobins. • The assembly characteristics of SLH4 and SC16 are distinct, outlining how surface properties of hydrophobins influence their function.
Organizational Affiliation:
Department of Biochemistry & Molecular Biology, Dalhousie University, Halifax, NS, Canada.