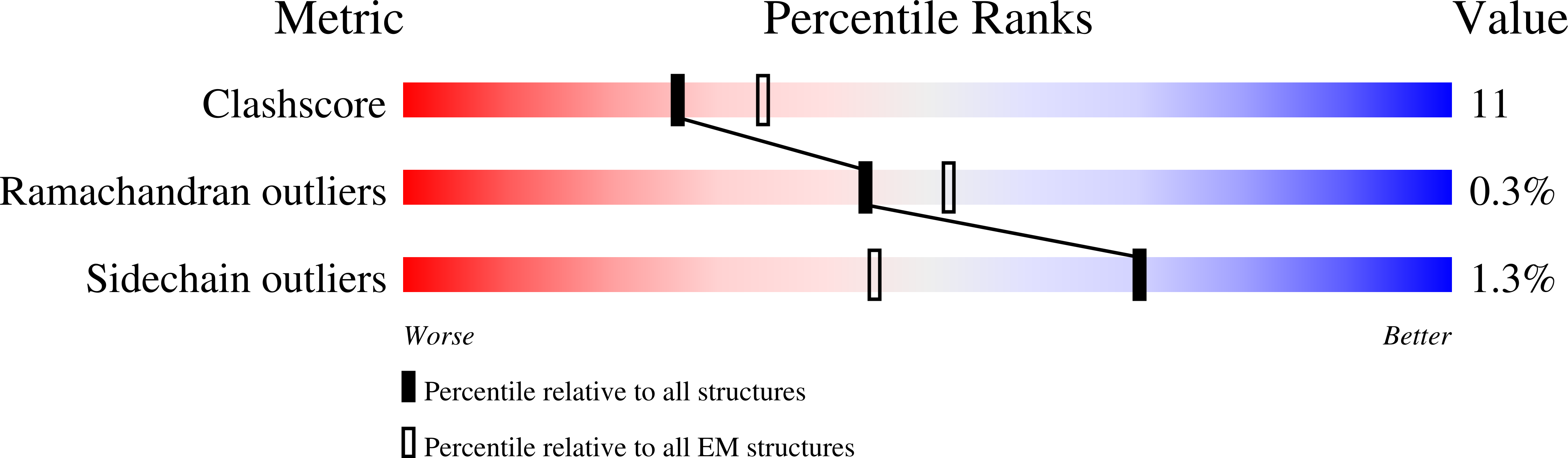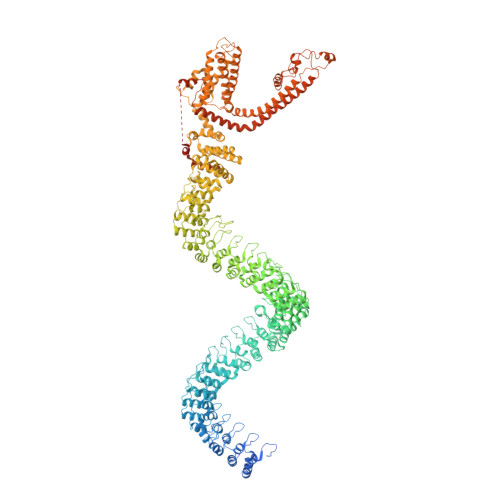Electron cryo-microscopy structure of the mechanotransduction channel NOMPC.
Jin, P., Bulkley, D., Guo, Y., Zhang, W., Guo, Z., Huynh, W., Wu, S., Meltzer, S., Cheng, T., Jan, L.Y., Jan, Y.N., Cheng, Y.(2017) Nature 547: 118-122
- PubMed: 28658211
- DOI: https://doi.org/10.1038/nature22981
- Primary Citation of Related Structures:
5VKQ - PubMed Abstract:
Mechanosensory transduction for senses such as proprioception, touch, balance, acceleration, hearing and pain relies on mechanotransduction channels, which convert mechanical stimuli into electrical signals in specialized sensory cells. How force gates mechanotransduction channels is a central question in the field, for which there are two major models. One is the membrane-tension model: force applied to the membrane generates a change in membrane tension that is sufficient to gate the channel, as in the bacterial MscL channel and certain eukaryotic potassium channels. The other is the tether model: force is transmitted via a tether to gate the channel. The transient receptor potential (TRP) channel NOMPC is important for mechanosensation-related behaviours such as locomotion, touch and sound sensation across different species including Caenorhabditis elegans, Drosophila and zebrafish. NOMPC is the founding member of the TRPN subfamily, and is thought to be gated by tethering of its ankyrin repeat domain to microtubules of the cytoskeleton. Thus, a goal of studying NOMPC is to reveal the underlying mechanism of force-induced gating, which could serve as a paradigm of the tether model. NOMPC fulfils all the criteria that apply to mechanotransduction channels and has 29 ankyrin repeats, the largest number among TRP channels. A key question is how the long ankyrin repeat domain is organized as a tether that can trigger channel gating. Here we present a de novo atomic structure of Drosophila NOMPC determined by single-particle electron cryo-microscopy. Structural analysis suggests that the ankyrin repeat domain of NOMPC resembles a helical spring, suggesting its role of linking mechanical displacement of the cytoskeleton to the opening of the channel. The NOMPC architecture underscores the basis of translating mechanical force into an electrical signal within a cell.
Organizational Affiliation:
Department of Physiology, University of California, San Francisco, California 94158, USA.