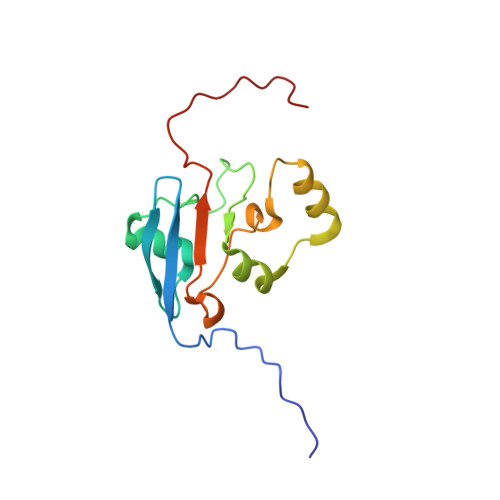
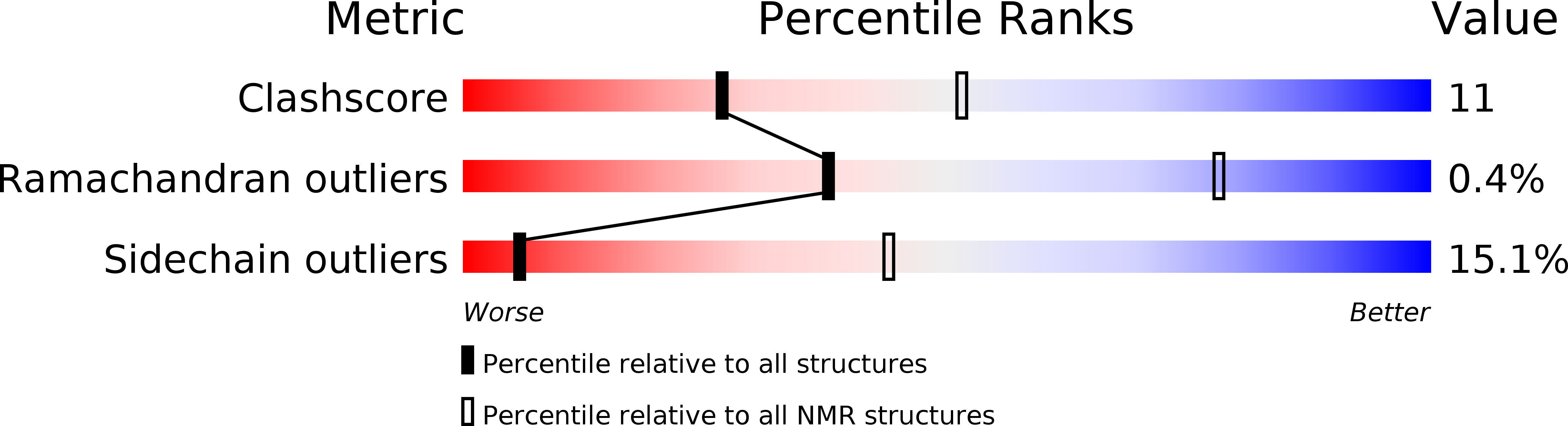Solution structure for an Encephalitozoon cuniculi adrenodoxin-like protein in the oxidized state.
Shaheen, S., Barrett, K.F., Subramanian, S., Arnold, S.L.M., Laureanti, J.A., Myler, P.J., Van Voorhis, W.C., Buchko, G.W.(2020) Protein Sci 29: 809-817
- PubMed: 31912584
- DOI: https://doi.org/10.1002/pro.3818
- Primary Citation of Related Structures:
5UJ5 - PubMed Abstract:
Encephalitozoon cuniculi is a unicellular, obligate intracellular eukaryotic parasite in the Microsporidia family and one of the agents responsible for microsporidosis infections in humans. Like most Microsporidia, the genome of E. cuniculi is markedly reduced and the organism contains mitochondria-like organelles called mitosomes instead of mitochondria. Here we report the solution NMR structure for a protein physically associated with mitosome-like organelles in E. cuniculi, the 128-residue, adrenodoxin-like protein Ec-Adx (UniProt ID Q8SV19) in the [2Fe-2S] ferredoxin superfamily. Oxidized Ec-Adx contains a mixed four-strand β-sheet, β2-β1-β4-β3 (↓↑↑↓), loosely encircled by three α-helices and two 3 10 -helices. This fold is similar to the structure observed in other adrenodoxin and adrenodoxin-like proteins except for the absence of a fifth anti-parallel β-strand next to β3 and the position of α3. Cross peaks are missing or cannot be unambiguously assigned for 20 amide resonances in the 1 H- 15 N HSQC spectrum of Ec-Adx. These missing residues are clustered primarily in two regions, G48-V61 and L94-L98, containing the four cysteine residues predicted to ligate the paramagnetic [2Fe-2S] cluster. Missing amide resonances in 1 H- 15 N HSQC spectra are detrimental to NMR-based solution structure calculations because 1 H- 1 H NOE restraints are absent (glass half-empty) and this may account for the absent β-strand (β5) and the position of α3 in oxidized Ec-Adx. On the other hand, the missing amide resonances unambiguously identify the presence, and immediate environment, of the paramagnetic [2Fe-2S] cluster in oxidized Ec-Adx (glass half-full).
Organizational Affiliation:
Department of Medicine, Division of Allergy and Infectious Disease, Center for Emerging and Re-emerging Infectious Disease, University of Washington, Seattle, Washington.