Structural Analysis Reveals the Deleterious Effects of Telomerase Mutations in Bone Marrow Failure Syndromes.
Hoffman, H., Rice, C., Skordalakes, E.(2017) J Biol Chem 292: 4593-4601
- PubMed: 28154186
- DOI: https://doi.org/10.1074/jbc.M116.771204
- Primary Citation of Related Structures:
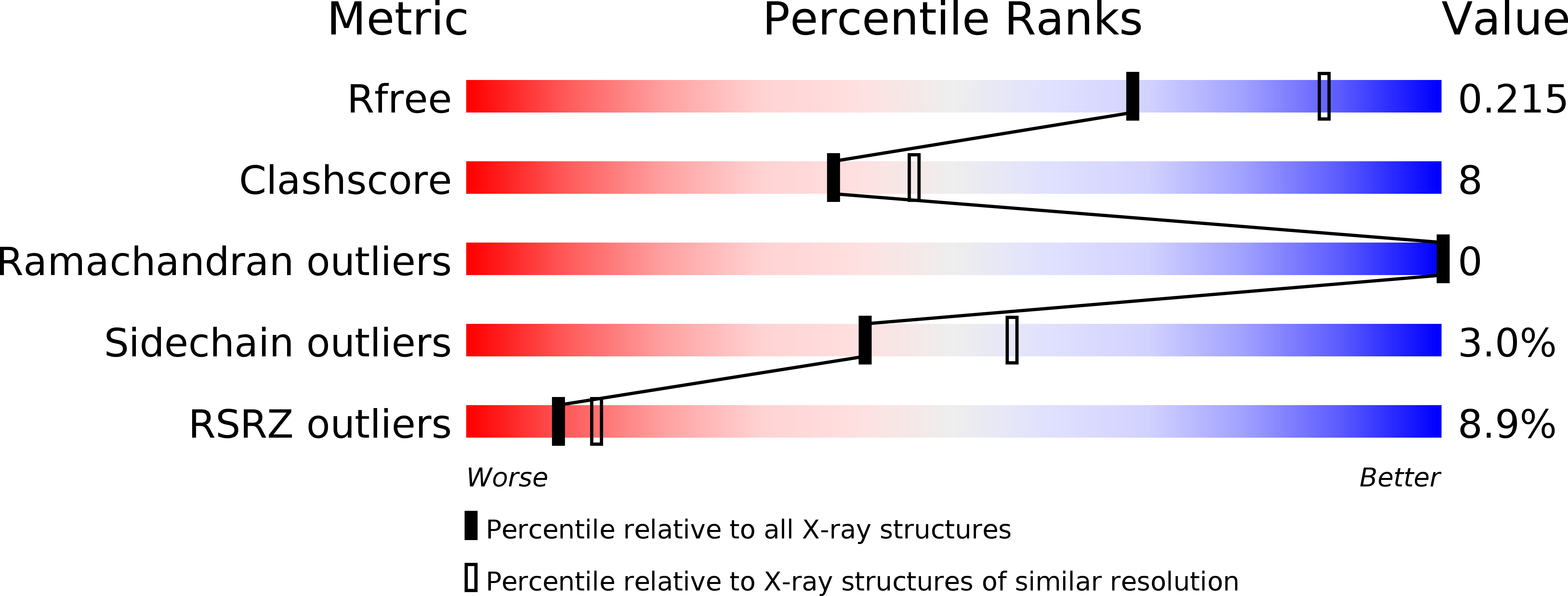
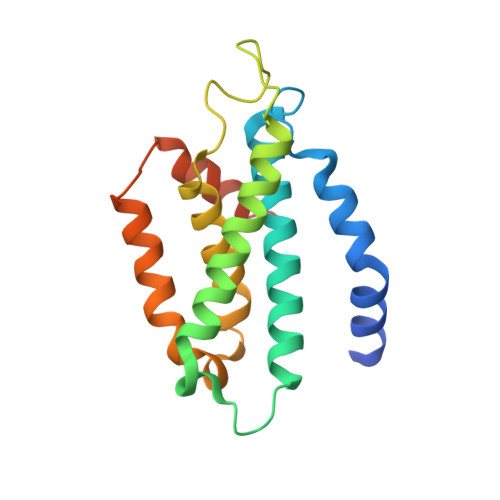
5UGW - PubMed Abstract:
Naturally occurring mutations in the ribonucleoprotein reverse transcriptase, telomerase, are associated with the bone marrow failure syndromes dyskeratosis congenita, aplastic anemia, and idiopathic pulmonary fibrosis. However, the mechanism by which these mutations impact telomerase function remains unknown. Here we present the structure of the human telomerase C-terminal extension (or thumb domain) determined by the method of single-wavelength anomalous diffraction to 2.31 Å resolution. We also used direct telomerase activity and nucleic acid binding assays to explain how naturally occurring mutations within this portion of telomerase contribute to human disease. The single mutations localize within three highly conserved regions of the telomerase thumb domain referred to as motifs E-I (thumb loop and helix), E-II, and E-III (the FVYL pocket, comprising the hydrophobic residues Phe-1012, Val-1025, Tyr-1089, and Leu-1092). Biochemical data show that the mutations associated with dyskeratosis congenita, aplastic anemia, and idiopathic pulmonary fibrosis disrupt the binding between the protein subunit reverse transcriptase of the telomerase and its nucleic acid substrates leading to loss of telomerase activity and processivity. Collectively our data show that although these mutations do not alter the overall stability or expression of telomerase reverse transcriptase, these rare genetic disorders are associated with an impaired telomerase holoenzyme that is unable to correctly assemble with its nucleic acid substrates, leading to incomplete telomere extension and telomere attrition, which are hallmarks of these diseases.
Organizational Affiliation:
From the Department of Gene Expression and Regulation, Wistar Institute, Philadelphia, Pennsylvania 19104 and.