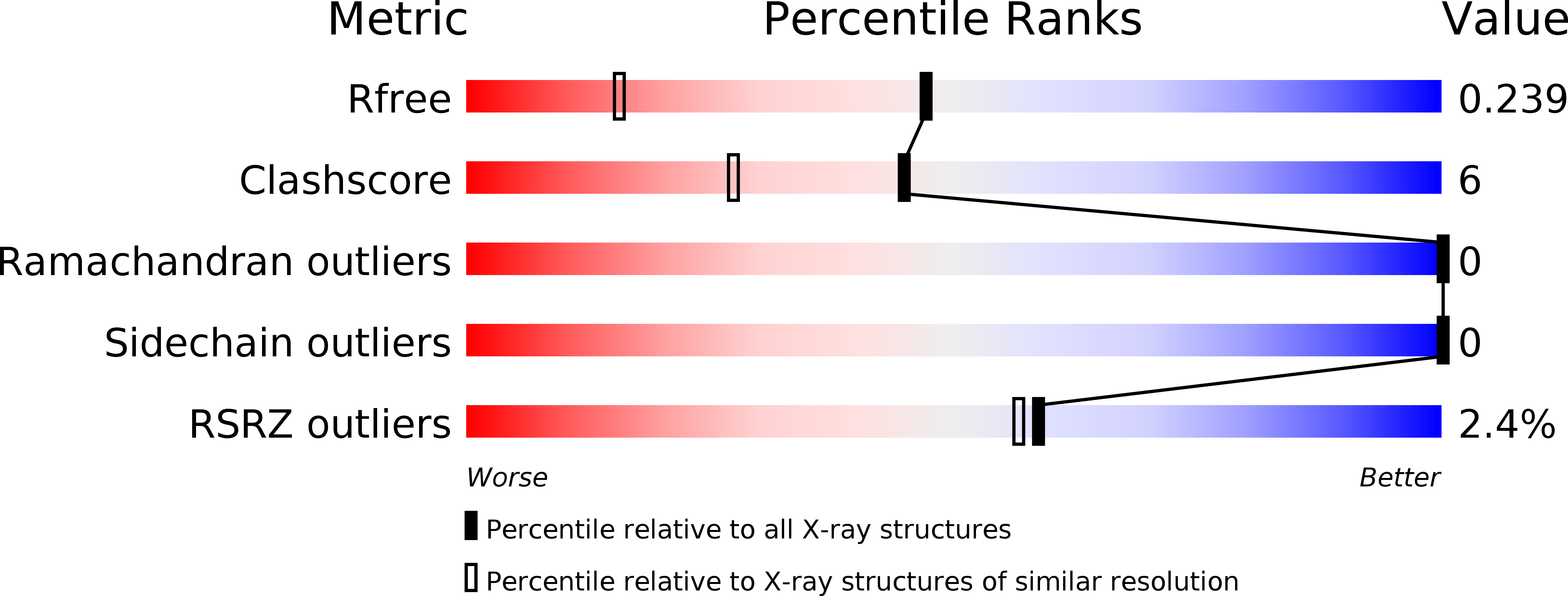
Recognition of Histone H3K14 Acylation by MORF.
Klein, B.J., Simithy, J., Wang, X., Ahn, J., Andrews, F.H., Zhang, Y., Cote, J., Shi, X., Garcia, B.A., Kutateladze, T.G.(2017) Structure 25: 650-654.e2
- PubMed: 28286003
- DOI: https://doi.org/10.1016/j.str.2017.02.003
- Primary Citation of Related Structures:
5U2J - PubMed Abstract:
The monocytic leukemia zinc-finger protein-related factor (MORF) is a transcriptional coactivator and a catalytic subunit of the lysine acetyltransferase complex implicated in cancer and developmental diseases. We have previously shown that the double plant homeodomain finger (DPF) of MORF is capable of binding to acetylated histone H3. Here we demonstrate that the DPF of MORF recognizes many newly identified acylation marks. The mass spectrometry study provides comprehensive analysis of H3K14 acylation states in vitro and in vivo. The crystal structure of the MORF DPF-H3K14butyryl complex offers insight into the selectivity of this reader toward lipophilic acyllysine substrates. Together, our findings support the mechanism by which the acetyltransferase MORF promotes spreading of histone acylation.
Organizational Affiliation:
Department of Pharmacology, University of Colorado School of Medicine, Aurora, CO 80045, USA.