Structure and Function of the RING Domains of RNF20 and RNF40, Dimeric E3 Ligases that Monoubiquitylate Histone H2B.
Foglizzo, M., Middleton, A.J., Day, C.L.(2016) J Mol Biol 428: 4073-4086
- PubMed: 27569044
- DOI: https://doi.org/10.1016/j.jmb.2016.07.025
- Primary Citation of Related Structures:
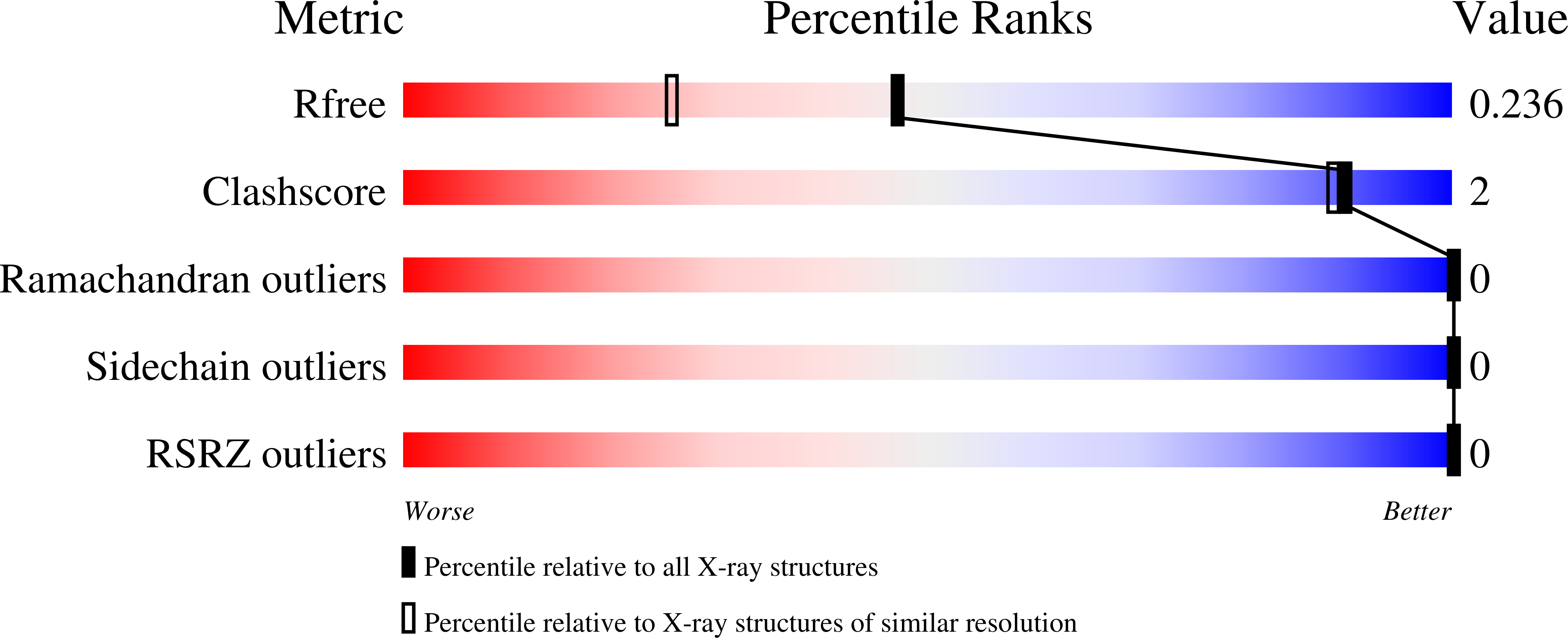
5TRB - PubMed Abstract:
Monoubiquitylation of histone H2B is a post-translational mark that plays key roles in regulation of transcription and genome stability. In humans, attachment of ubiquitin to lysine 120 of histone H2B depends on the activity of the E2 ubiquitin-conjugating enzyme, Ube2B, and the really interesting new gene (RING) E3 ligases, RING finger protein (RNF) 20 and RNF40. To better understand the molecular basis of this modification, we have solved the crystal structure of the RNF20 RING domain and show that it is a homodimer that specifically interacts with the Ube2B~Ub conjugate. By mutating residues at the E3-E2 and E3-ubiquitin interfaces, we identify key contacts required for interaction of the RNF20 RING domain with the Ube2B~Ub conjugate. These mutants were used to generate a structure-based model of the RNF20-Ube2B~Ub complex that reveals differences from other RING-E2~Ub complexes, and suggests how the RNF20-Ube2B~Ub complex might interact with its nucleosomal substrate. Additionally, we show that the RING domains of RNF20 and RNF40 can form a stable heterodimer that is active. Together, our studies provide new insights into the mechanisms that regulate RNF20-mediated ubiquitin transfer from Ube2B.
Organizational Affiliation:
Biochemistry Department, Otago School of Medical Sciences, University of Otago, Dunedin 9054, New Zealand.