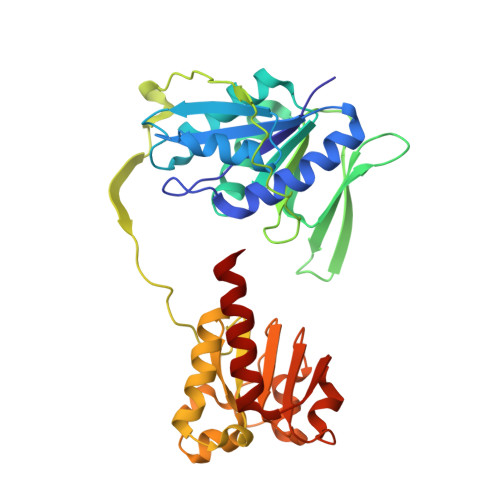
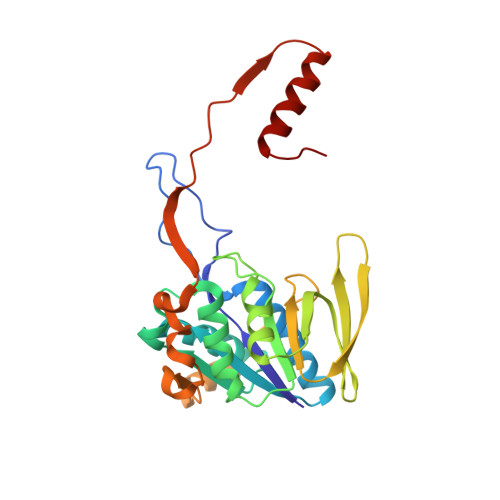
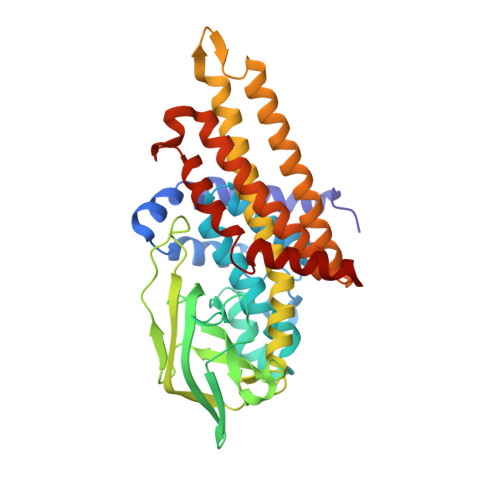
The semiquinone swing in the bifurcating electron transferring flavoprotein/butyryl-CoA dehydrogenase complex from Clostridium difficile.
Demmer, J.K., Pal Chowdhury, N., Selmer, T., Ermler, U., Buckel, W.(2017) Nat Commun 8: 1577-1577
- PubMed: 29146947
- DOI: https://doi.org/10.1038/s41467-017-01746-3
- Primary Citation of Related Structures:
5OL2 - PubMed Abstract:
The electron transferring flavoprotein/butyryl-CoA dehydrogenase (EtfAB/Bcd) catalyzes the reduction of one crotonyl-CoA and two ferredoxins by two NADH within a flavin-based electron-bifurcating process. Here we report on the X-ray structure of the Clostridium difficile (EtfAB/Bcd) 4 complex in the dehydrogenase-conducting D-state, α-FAD (bound to domain II of EtfA) and δ-FAD (bound to Bcd) being 8 Å apart. Superimposing Acidaminococcus fermentans EtfAB onto C. difficile EtfAB/Bcd reveals a rotation of domain II of nearly 80°. Further rotation by 10° brings EtfAB into the bifurcating B-state, α-FAD and β-FAD (bound to EtfB) being 14 Å apart. This dual binding mode of domain II, substantiated by mutational studies, resembles findings in non-bifurcating EtfAB/acyl-CoA dehydrogenase complexes. In our proposed mechanism, NADH reduces β-FAD, which bifurcates. One electron goes to ferredoxin and one to α-FAD, which swings over to reduce δ-FAD to the semiquinone. Repetition affords a second reduced ferredoxin and δ-FADH - , which reduces crotonyl-CoA.
Organizational Affiliation:
Max-Planck-Institut für Biophysik, Max-von-Laue-Str. 3, 60438, Frankfurt am Main, Germany.