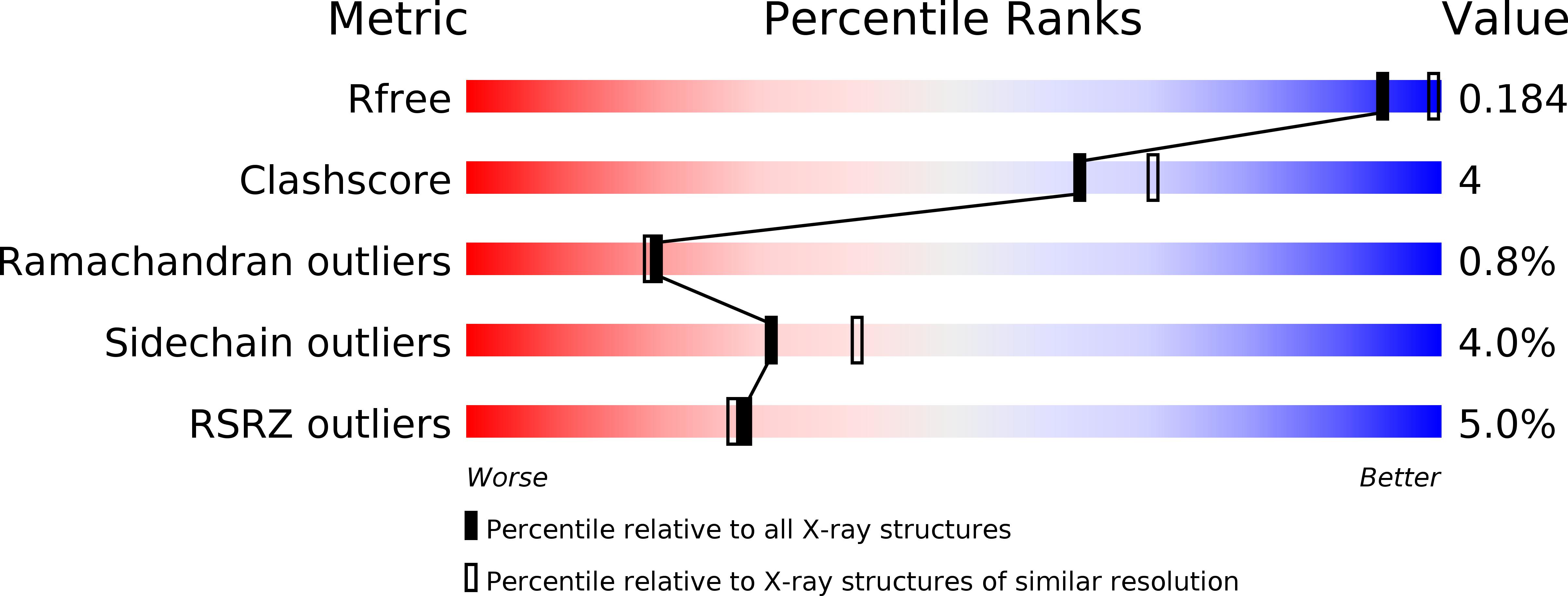
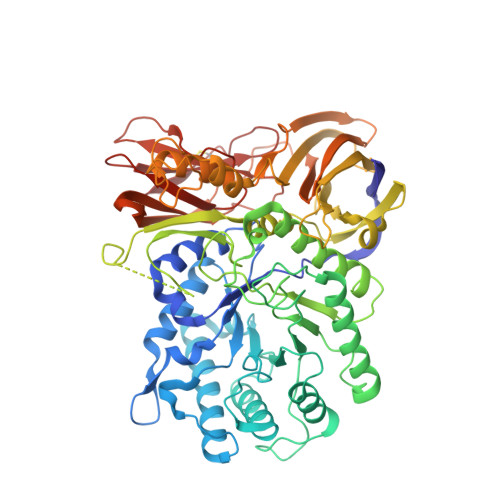
The bacterial orthologue of Human a-L-iduronidase does not need N-glycan post-translational modifications to be catalytically competent: Crystallography and QM/MM insights into Mucopolysaccharidosis I.
Raich, L., Valero-Gonzalez, J., Castro-Lopez, J., Millan, C., Jimenez-Garcia, M.J., Nieto, P., Uson, I., Hurtado-Guerrero, R., Rovira, C.To be published.