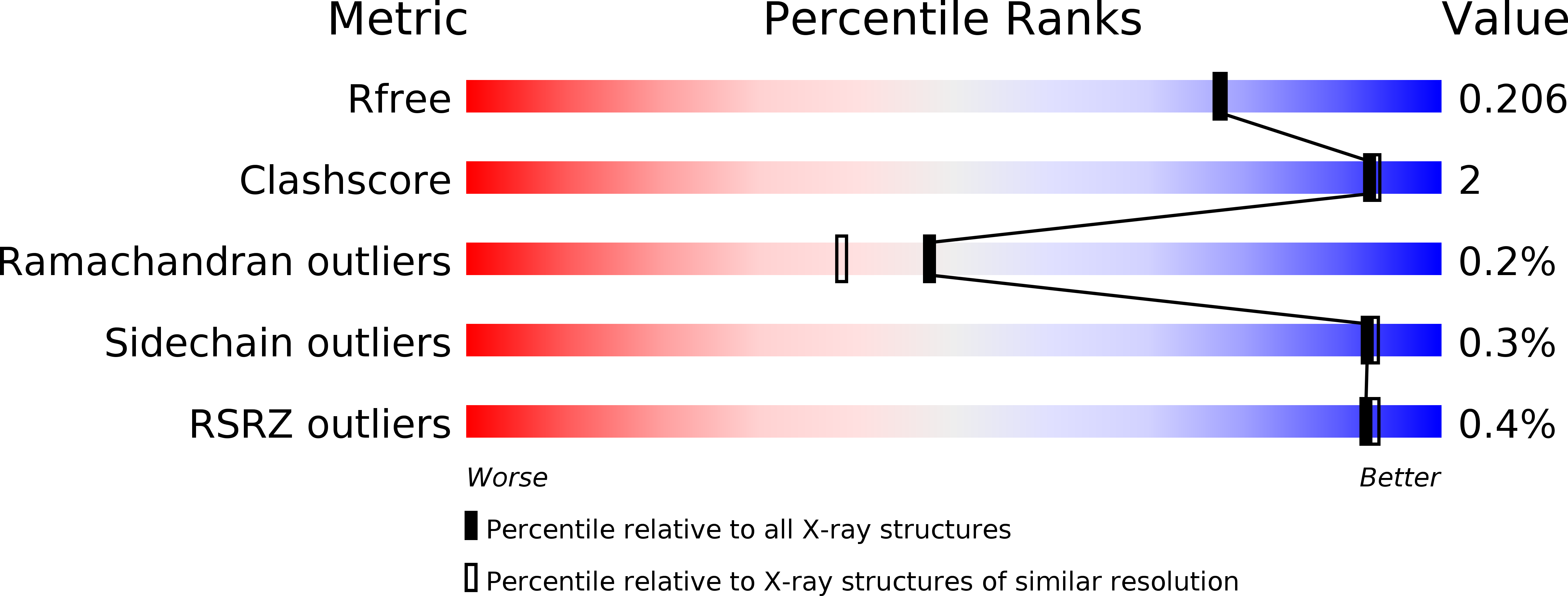
Structural and Functional Basis of C-Methylation of Coumarin Scaffolds by NovO.
Sadler, J.C., Chung, C.H., Mosley, J.E., Burley, G.A., Humphreys, L.D.(2017) ACS Chem Biol 12: 374-379
- PubMed: 28068060
- DOI: https://doi.org/10.1021/acschembio.6b01053
- Primary Citation of Related Structures:
5MGZ - PubMed Abstract:
C-methylation of aromatic small molecules by C-methyltransferases (C-MTs) is an important biological transformation that involves C-C bond formation using S-adenosyl-l-methionine (SAM) as the methyl donor. Here, two advances in the mechanistic understanding of C-methylation of the 8-position of coumarin substrates catalyzed by the C-MT NovO from Streptomyces spheroides are described. First, a crystal structure of NovO reveals the Arg116-Asn117 and His120-Arg121 motifs are essential for coumarin substrate binding. Second, the active-site His120 is responsible for deprotonation of the phenolic 7-hydroxyl group on the coumarin substrate, activating the rate-determining methyl transfer step from SAM. This work expands our mechanistic knowledge of C-MTs, which could be used in the downstream development of engineered biocatalysts for small molecule C-alkylations.
Organizational Affiliation:
GlaxoSmithKline Medicines Research Centre , Gunnels Wood Road, Stevenage, SG1 2NY , United Kingdom.