Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies.
Hofbauer, S., Mlynek, G., Milazzo, L., Puhringer, D., Maresch, D., Schaffner, I., Furtmuller, P.G., Smulevich, G., Djinovic-Carugo, K., Obinger, C.(2016) FEBS J 283: 4386-4401
- PubMed: 27758026
- DOI: https://doi.org/10.1111/febs.13930
- Primary Citation of Related Structures:
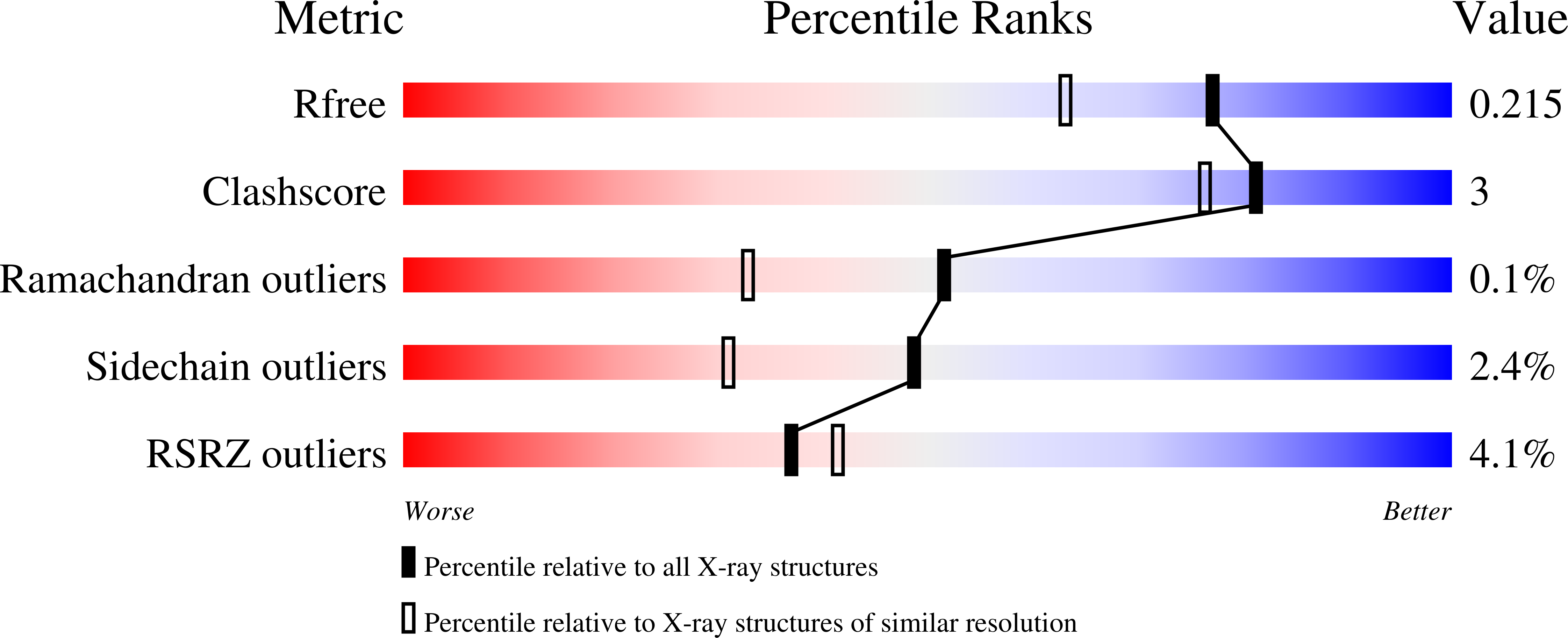
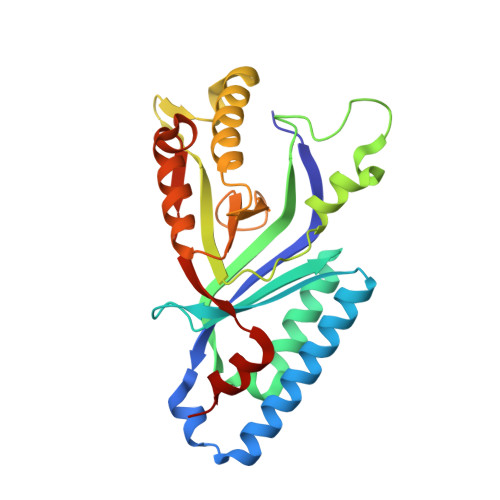
5LOQ - PubMed Abstract:
Heme biosynthesis in Gram-positive bacteria follows a recently described coproporphyrin-dependent pathway with HemQ catalyzing the decarboxylation of coproheme to heme b. Here we present the first crystal structure of a HemQ (homopentameric coproheme-HemQ from Listeria monocytogenes) at 1.69 Å resolution and the conversion of coproheme to heme b followed by UV-vis and resonance Raman spectroscopy as well as mass spectrometry. The ferric five-coordinated coproheme iron of HemQ is weakly bound by a neutral proximal histidine H174. In the crystal structure of the resting state, the distal Q187 (conserved in Firmicutes HemQ) is H-bonded with propionate p2 and the hydrophobic distal cavity lacks solvent water molecules. Two H 2 O 2 molecules are shown to be necessary for decarboxylation of the propionates p2 and p4, thereby forming the corresponding vinyl groups of heme b. The overall reaction is relatively slow (k cat /K M = 1.8 × 10 2 m -1 ·s -1 at pH 7.0) and occurs in a stepwise manner with a three-propionate intermediate. We present the noncovalent interactions between coproheme and the protein and propose a two-step reaction mechanism. Furthermore, the structure of coproheme-HemQ is compared to that of the phylogenetically related heme b-containing chlorite dismutases.
Organizational Affiliation:
Department for Structural and Computational Biology, Max F. Perutz Laboratories, University of Vienna, Austria.