Cytochrome c Can Form a Well-Defined Binding Pocket for Hydrocarbons.
McClelland, L.J., Steele, H.B., Whitby, F.G., Mou, T.C., Holley, D., Ross, J.B., Sprang, S.R., Bowler, B.E.(2016) J Am Chem Soc 138: 16770-16778
- PubMed: 27990813
- DOI: https://doi.org/10.1021/jacs.6b10745
- Primary Citation of Related Structures:
5KKE, 5KLU, 5T7H - PubMed Abstract:
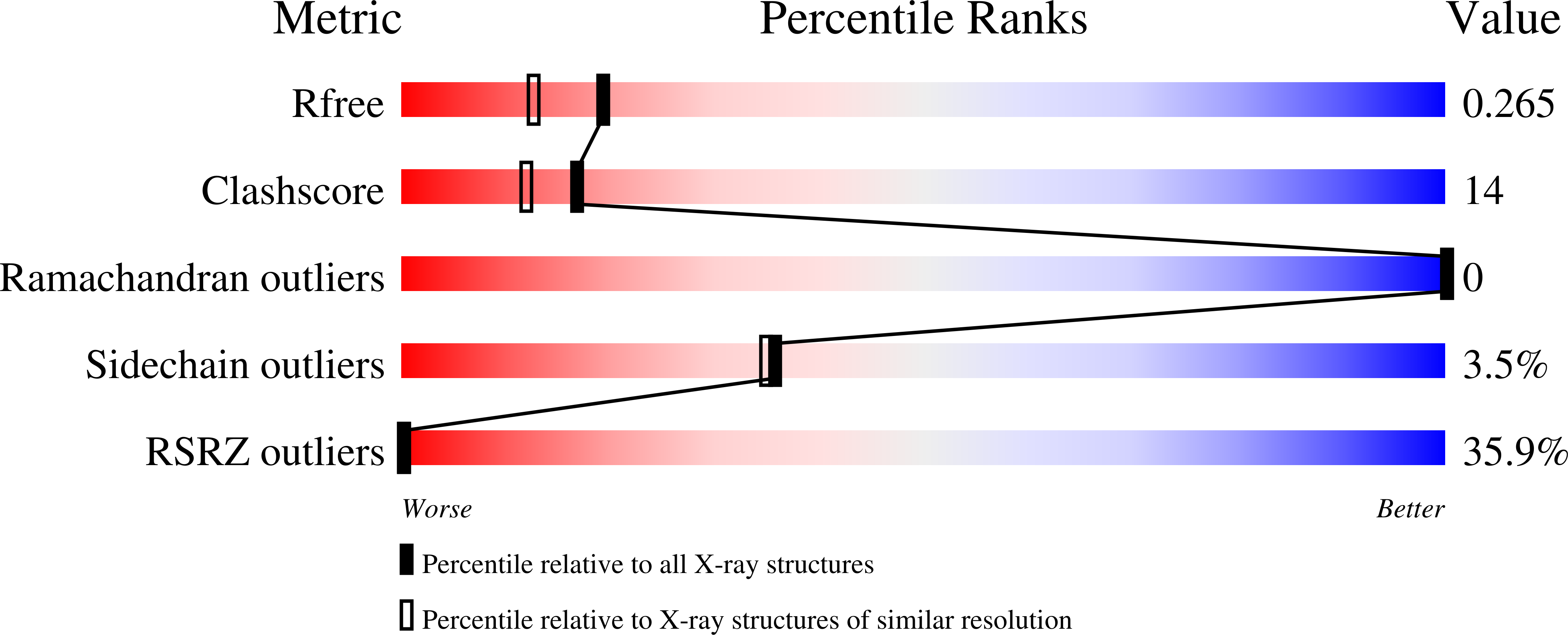
Cytochrome c can acquire peroxidase activity when it binds to cardiolipin in mitochondrial membranes. The resulting oxygenation of cardiolipin by cytochrome c provides an early signal for the onset of apoptosis. The structure of this enzyme-substrate complex is a matter of considerable debate. We present three structures at 1.7-2.0 Å resolution of a domain-swapped dimer of yeast iso-1-cytochrome c with the detergents, CYMAL-5, CYMAL-6, and ω-undecylenyl-β-d-maltopyranoside, bound in a channel that places the hydrocarbon moieties of these detergents next to the heme. The heme is poised for peroxidase activity with water bound in place of Met80, which serves as the axial heme ligand when cytochrome c functions as an electron carrier. The hydroxyl group of Tyr67 sits 3.6-4.0 Å from the nearest carbon of the detergents, positioned to act as a relay in radical abstraction during peroxidase activity. Docking studies with linoleic acid, the most common fatty acid component of cardiolipin, show that C11 of linoleic acid can sit adjacent to Tyr67 and the heme, consistent with the oxygenation pattern observed in lipidomics studies. The well-defined hydrocarbon binding pocket provides atomic resolution evidence for the extended lipid anchorage model for cytochrome c/cardiolipin binding. Dimer dissociation/association kinetics for yeast versus equine cytochrome c indicate that formation of mammalian cytochrome c dimers in vivo would require catalysis. However, the dimer structure shows that only a modest deformation of monomeric cytochrome c would suffice to form the hydrocarbon binding site occupied by these detergents.
Organizational Affiliation:
Department of Chemistry & Biochemistry, University of Montana , Missoula, Montana 59812, United States.