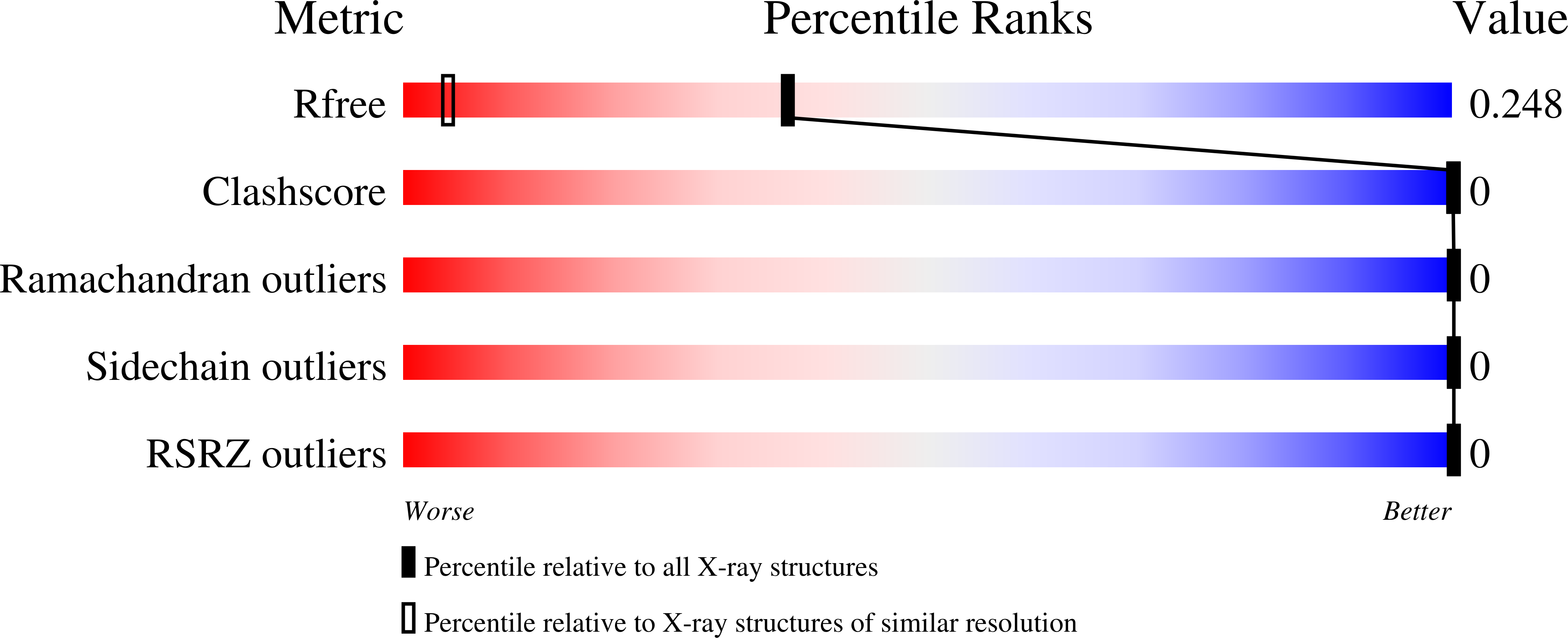
XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction.
Thomaston, J.L., Woldeyes, R.A., Nakane, T., Yamashita, A., Tanaka, T., Koiwai, K., Brewster, A.S., Barad, B.A., Chen, Y., Lemmin, T., Uervirojnangkoorn, M., Arima, T., Kobayashi, J., Masuda, T., Suzuki, M., Sugahara, M., Sauter, N.K., Tanaka, R., Nureki, O., Tono, K., Joti, Y., Nango, E., Iwata, S., Yumoto, F., Fraser, J.S., DeGrado, W.F.(2017) Proc Natl Acad Sci U S A 114: 13357-13362
- PubMed: 28835537
- DOI: https://doi.org/10.1073/pnas.1705624114
- Primary Citation of Related Structures:
5JOO, 5TTC, 5UM1 - PubMed Abstract:
The M2 proton channel of influenza A is a drug target that is essential for the reproduction of the flu virus. It is also a model system for the study of selective, unidirectional proton transport across a membrane. Ordered water molecules arranged in "wires" inside the channel pore have been proposed to play a role in both the conduction of protons to the four gating His37 residues and the stabilization of multiple positive charges within the channel. To visualize the solvent in the pore of the channel at room temperature while minimizing the effects of radiation damage, data were collected to a resolution of 1.4 Å using an X-ray free-electron laser (XFEL) at three different pH conditions: pH 5.5, pH 6.5, and pH 8.0. Data were collected on the Inward open state, which is an intermediate that accumulates at high protonation of the His37 tetrad. At pH 5.5, a continuous hydrogen-bonded network of water molecules spans the vertical length of the channel, consistent with a Grotthuss mechanism model for proton transport to the His37 tetrad. This ordered solvent at pH 5.5 could act to stabilize the positive charges that build up on the gating His37 tetrad during the proton conduction cycle. The number of ordered pore waters decreases at pH 6.5 and 8.0, where the Inward open state is less stable. These studies provide a graphical view of the response of water to a change in charge within a restricted channel environment.
Organizational Affiliation:
Department of Pharmaceutical Chemistry, University of California, San Francisco, CA 94158.