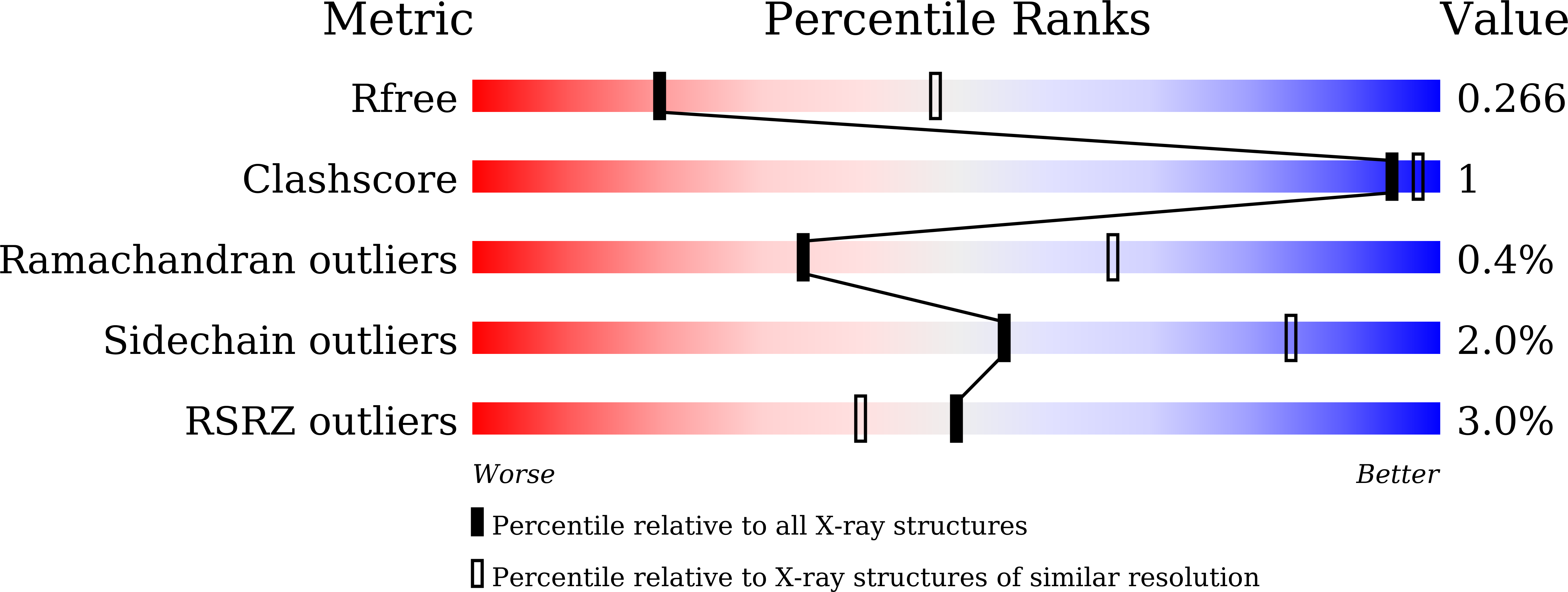
Crystal structures of arginine sensor CASTOR1 in arginine-bound and ligand free states
Zhou, Y., Wang, C., Xiao, Q., Guo, L.(2019) Biochem Biophys Res Commun 508: 387-391
- PubMed: 30503338
- DOI: https://doi.org/10.1016/j.bbrc.2018.11.147
- Primary Citation of Related Structures:
5GT7, 5GT8 - PubMed Abstract:
The mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) is a master regulator of metabolism and cell growth. Among the numerous extracellular and intracellular signals, certain amino acids activate mTORC1 in a Rag-dependent manner. Arginine can stimulate mTORC1 activity by releasing the inhibitor CASTOR1 (Cellular Arginine Sensor of mTORC1) from GATOR2, a positive regulator of mTORC1 which interacts with GATOR1, the GAP for RagA/B. Three groups have resolved the structures of arginine-CASTOR1 complex, shedding a new light on molecular basis of the regulation of mTORC1 activity by arginine. However, lacking the apo structure of CASTOR1 prelimited the molecular understanding of mechanism underlying mTORC1 regulation. Here, we report crystal structures of arginine sensor CASTOR1 in arginine-bound and ligand free states at 2.05 Å and 2.8 Å, respectively. Structural comparison of CASTOR1 between two states reveals near identical conformations, except in two loop regions. It indicates CASTOR1 does not undergo large conformational change during arginine binding. Therefore, we conclude a detailed structural interpretation of arginine sensing by CASTOR1 in mTORC1 pathway.
Organizational Affiliation:
State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, 610041, China.