An Atypical alpha / beta-Hydrolase Fold Revealed in the Crystal Structure of Pimeloyl-Acyl Carrier Protein Methyl Esterase BioG from Haemophilus influenzae
Shi, J., Cao, X., Chen, Y., Cronan, J.E., Guo, Z.(2016) Biochemistry 55: 6705-6717
- PubMed: 27933801
- DOI: https://doi.org/10.1021/acs.biochem.6b00818
- Primary Citation of Related Structures:
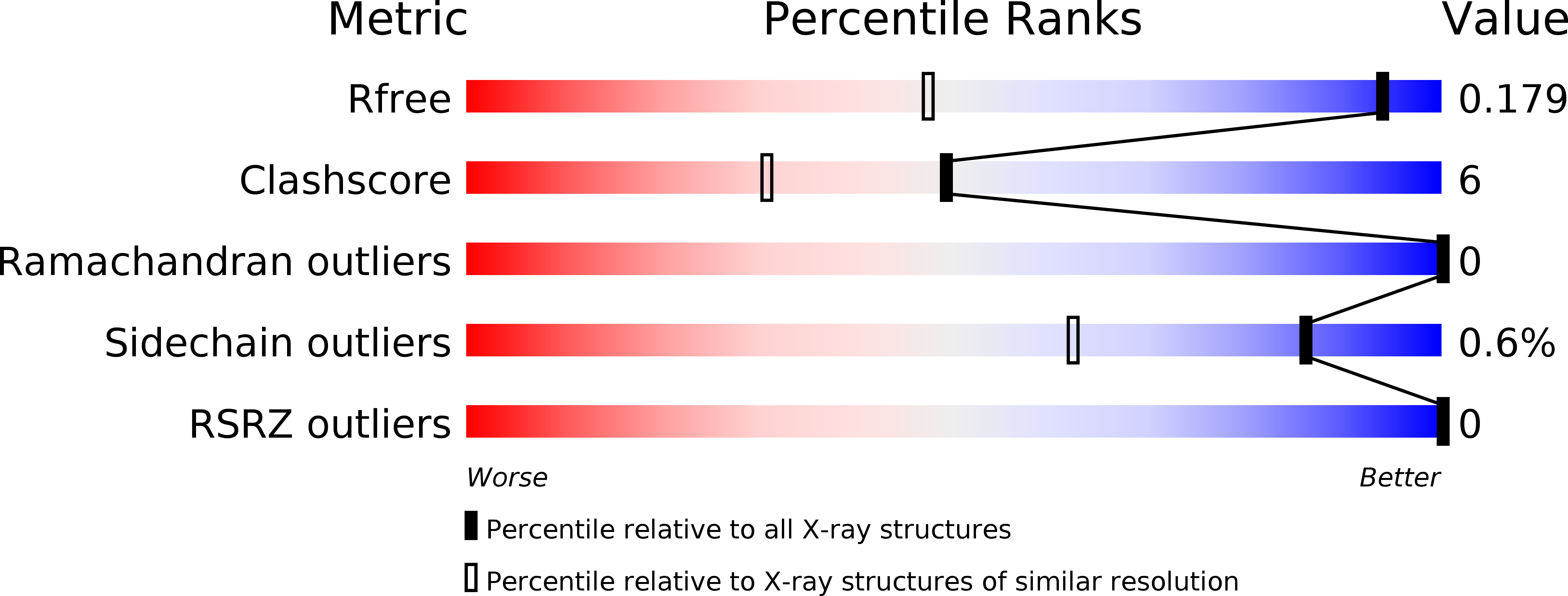
5GNG, 5H3B - PubMed Abstract:
Pimeloyl-acyl carrier protein (ACP) methyl esterase is an α/β-hydrolase that catalyzes the last biosynthetic step of pimeloyl-ACP, a key intermediate in biotin biosynthesis. Intriguingly, multiple nonhomologous isofunctional forms of this enzyme that lack significant sequence identity are present in diverse bacteria. One such esterase, Escherichia coli BioH, has been shown to be a typical α/β-hydrolase fold enzyme. To gain further insights into the role of this step in biotin biosynthesis, we have determined the crystal structure of another widely distributed pimeloyl-ACP methyl esterase, Haemophilus influenzae BioG, at 1.26 Å. The BioG structure is similar to the BioH structure and is composed of an α-helical lid domain and a core domain that contains a central seven-stranded β-pleated sheet. However, four of the six α-helices that flank both sides of the BioH core β-sheet are replaced with long loops in BioG, thus forming an unusual α/β-hydrolase fold. This structural variation results in a significantly decreased thermal stability of the enzyme. Nevertheless, the lid domain and the residues at the lid-core interface are well conserved between BioH and BioG, in which an analogous hydrophobic pocket for pimelate binding as well as similar ionic interactions with the ACP moiety are retained. Biochemical characterization of site-directed mutants of the residues hypothesized to interact with the ACP moiety supports a similar substrate interaction mode for the two enzymes. Consequently, these enzymes package the identical catalytic function under a considerably different protein surface.
Organizational Affiliation:
Department of Chemistry and State Key Lab for Molecular Neuroscience, The Hong Kong University of Science and Technology , Clear Water Bay, Kowloon, Hong Kong SAR, China.