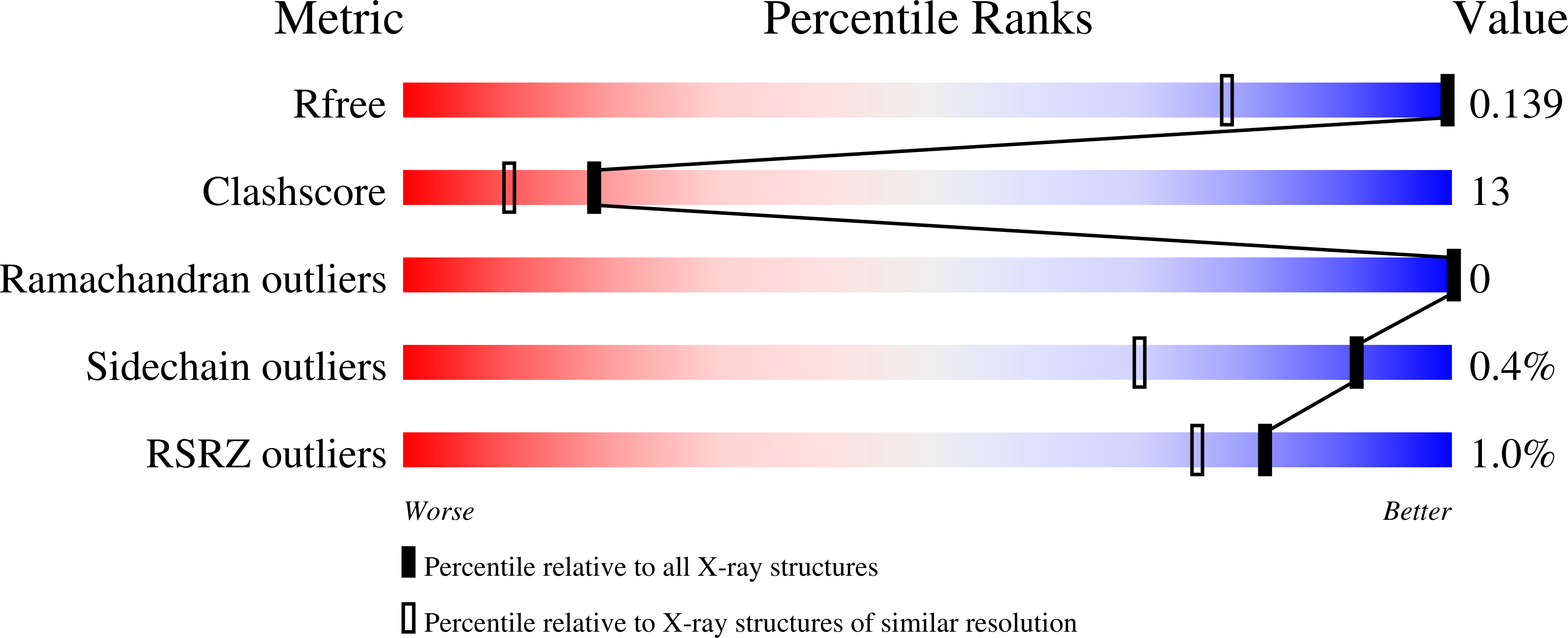
Evolutionary Analysis is a Powerful Complement to Energy Calculations Allowing Entropy-Driven Stabilization
Beerens, K., Mazurenko, S., Kunka, A., Marques, S.M., Hansen, N., Musil, M., Chaloupkova, R., Waterman, J., Brezovsky, J., Bednar, D., Prokop, Z., Damborsky, J.To be published.