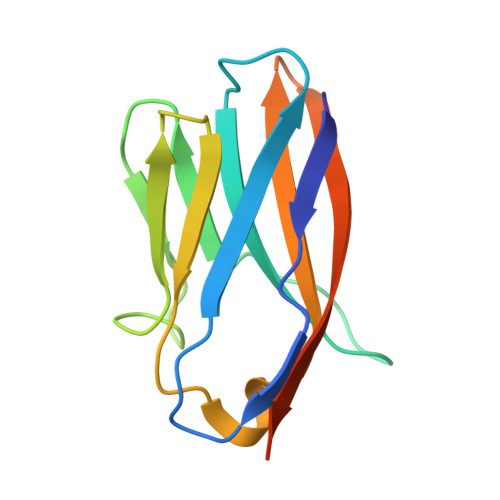
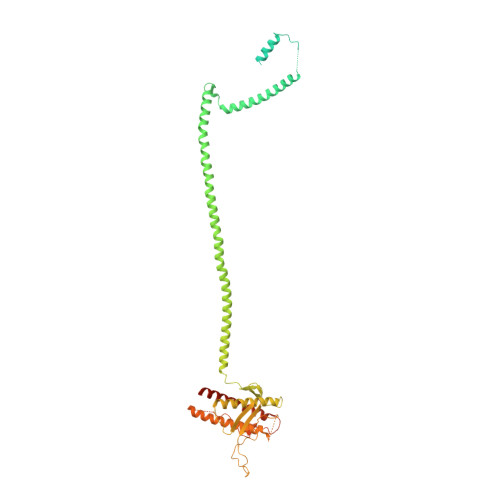
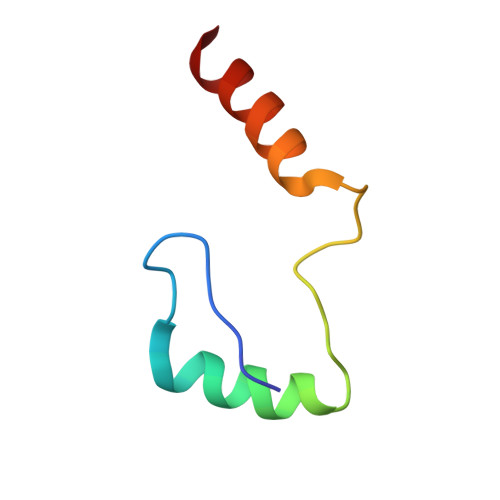
Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes.
Rostislavleva, K., Soler, N., Ohashi, Y., Zhang, L., Pardon, E., Burke, J.E., Masson, G.R., Johnson, C., Steyaert, J., Ktistakis, N.T., Williams, R.L.(2015) Science 350: aac7365-aac7365
- PubMed: 26450213
- DOI: https://doi.org/10.1126/science.aac7365
- Primary Citation of Related Structures:
5DFZ - PubMed Abstract:
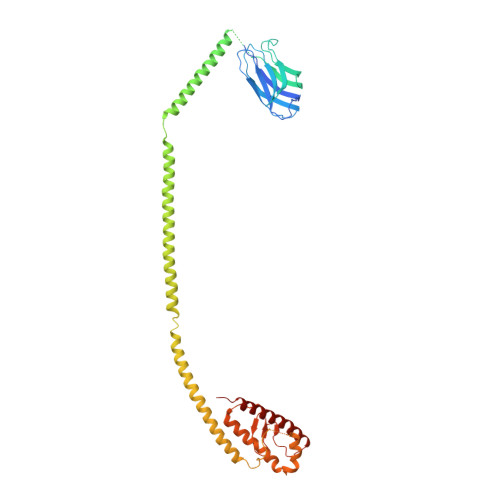
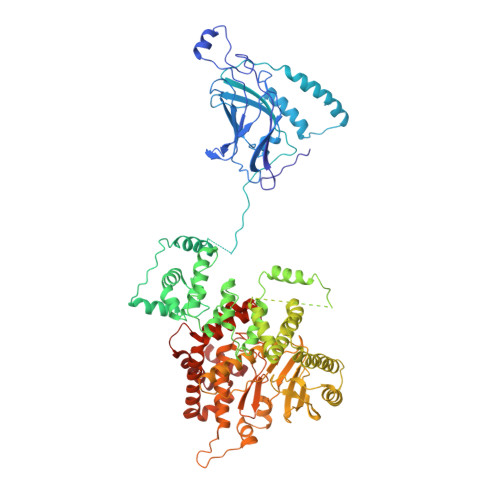
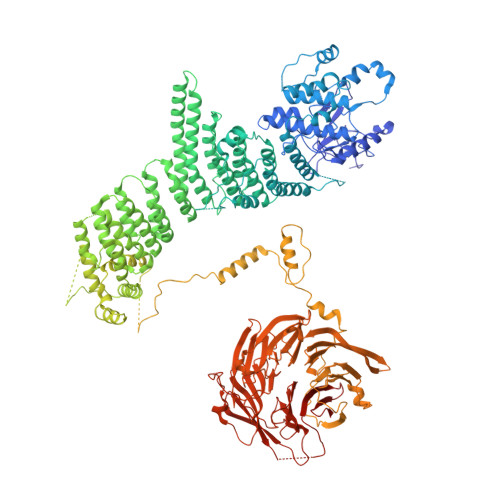
Phosphatidylinositol 3-kinase Vps34 complexes regulate intracellular membrane trafficking in endocytic sorting, cytokinesis, and autophagy. We present the 4.4 angstrom crystal structure of the 385-kilodalton endosomal complex II (PIK3C3-CII), consisting of Vps34, Vps15 (p150), Vps30/Atg6 (Beclin 1), and Vps38 (UVRAG). The subunits form a Y-shaped complex, centered on the Vps34 C2 domain. Vps34 and Vps15 intertwine in one arm, where the Vps15 kinase domain engages the Vps34 activation loop to regulate its activity. Vps30 and Vps38 form the other arm that brackets the Vps15/Vps34 heterodimer, suggesting a path for complex assembly. We used hydrogen-deuterium exchange mass spectrometry (HDX-MS) to reveal conformational changes accompanying membrane binding and identify a Vps30 loop that is critical for the ability of complex II to phosphorylate giant liposomes on which complex I is inactive.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Cambridge CB2 0QH, UK.