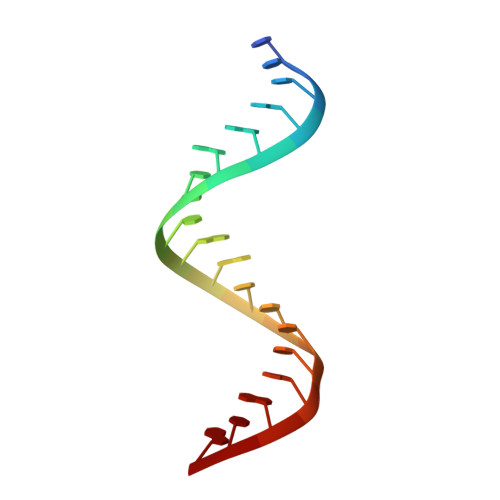
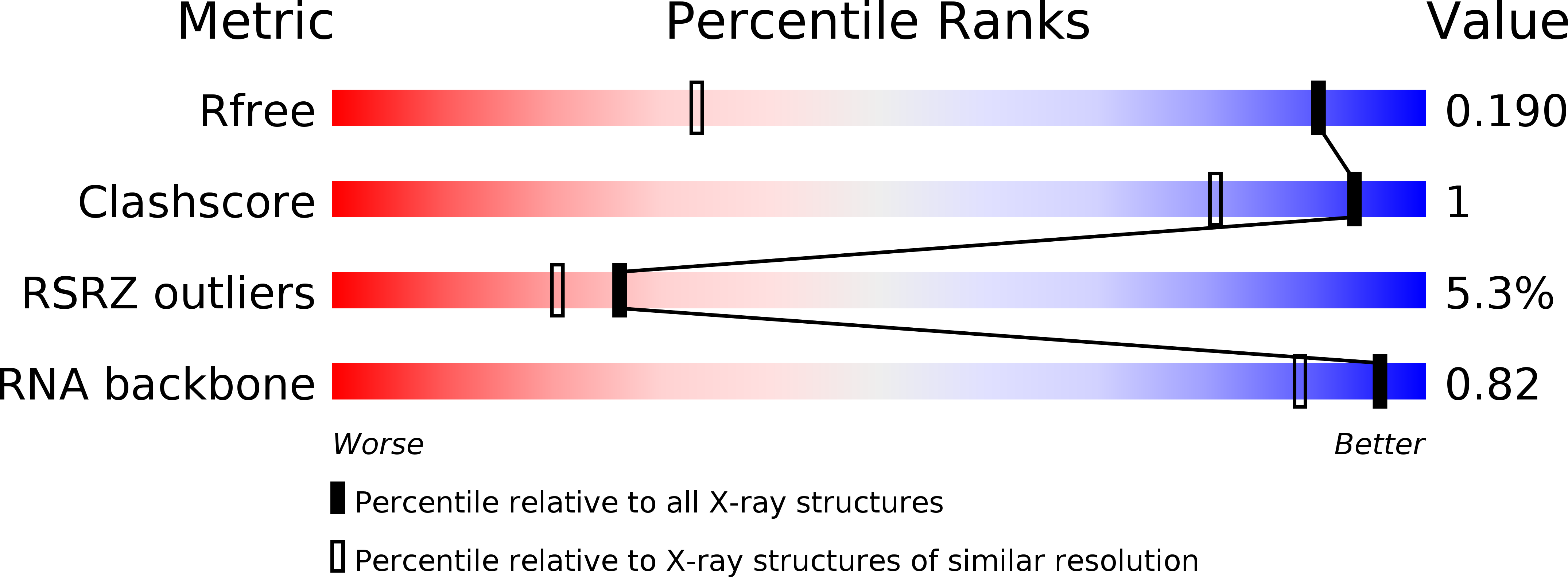1.25 angstrom resolution structure of an RNA 20-mer that binds to the TREX2 complex.
Valkov, E., Stewart, M.(2015) Acta Crystallogr F Struct Biol Commun 71: 1318-1321
- PubMed: 26457524
- DOI: https://doi.org/10.1107/S2053230X1501643X
- Primary Citation of Related Structures:
5C5W - PubMed Abstract:
The 1.25 Å resolution H32:R crystal structure of a 20 nt ribonucleotide that binds to the TREX-2 complex with high affinity shows a double-stranded RNA duplex arranged along a crystallographic 31 axis in which the antiparallel chains overlap by 18 nucleotides and are related by a crystallographic twofold axis. The duplex shows C-A, U-U and C-C noncanonical base pairings together with canonical Watson-Crick A-U and G-C pairs and a G-U wobble.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge Biomedical Campus, Cambridge CB2 0QH, England.