The Role of Lipids in Mechanosensation.
Pliotas, C., Dahl, A.C.E., Rasmussen, T., Mahendran, K.R., Smith, T.K., Marius, P., Gault, J., Banda, T., Rasmussen, A., Miller, S., Robinson, C.V., Bayley, H., Sansom, M.S.P., Booth, I.R., Naismith, J.H.(2015) Nat Struct Mol Biol 22: 991
- PubMed: 26551077
- DOI: https://doi.org/10.1038/nsmb.3120
- Primary Citation of Related Structures:
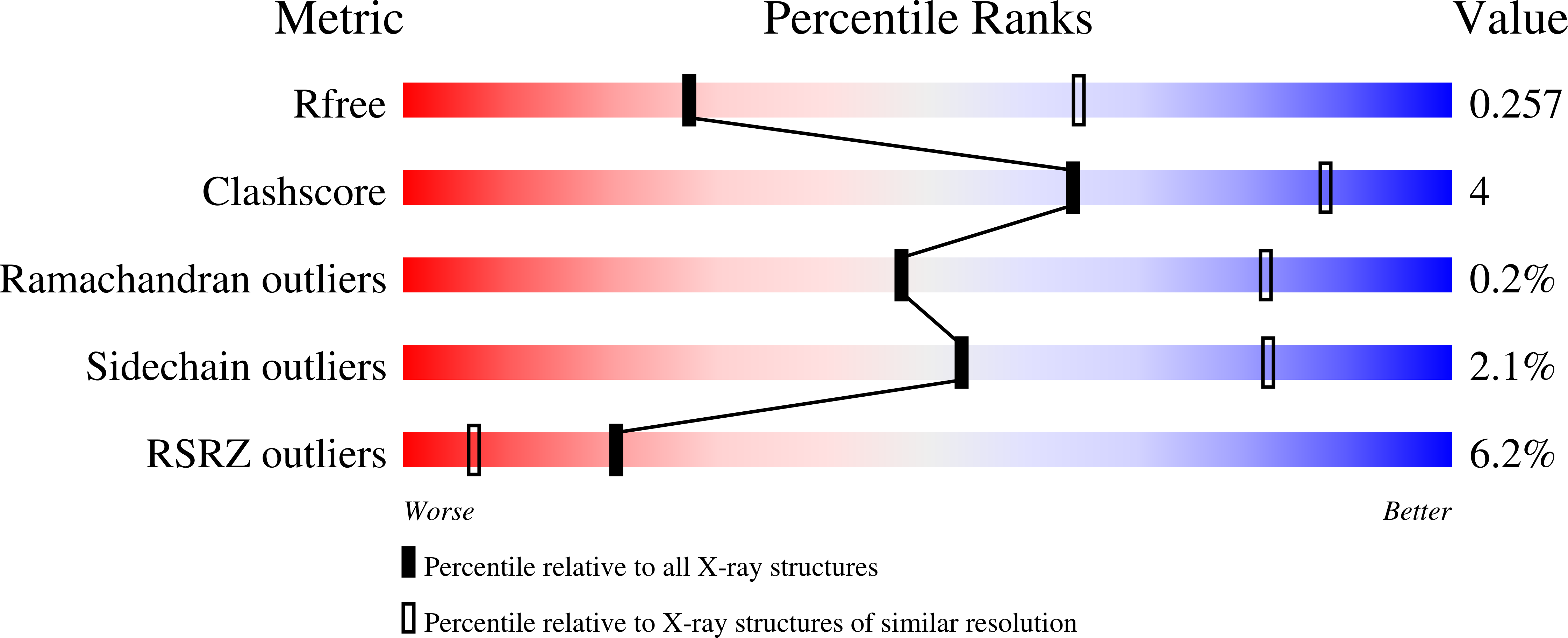
5AJI - PubMed Abstract:
The ability of proteins to sense membrane tension is pervasive in biology. A higher-resolution structure of the Escherichia coli small-conductance mechanosensitive channel MscS identifies alkyl chains inside pockets formed by the transmembrane helices (TMs). Purified MscS contains E. coli lipids, and fluorescence quenching demonstrates that phospholipid acyl chains exchange between bilayer and TM pockets. Molecular dynamics and biophysical analyses show that the volume of the pockets and thus the number of lipid acyl chains within them decreases upon channel opening. Phospholipids with one acyl chain per head group (lysolipids) displace normal phospholipids (with two acyl chains) from MscS pockets and trigger channel opening. We propose that the extent of acyl-chain interdigitation in these pockets determines the conformation of MscS. When interdigitation is perturbed by increased membrane tension or by lysolipids, the closed state becomes unstable, and the channel gates.
Organizational Affiliation:
Biomedical Sciences Research Complex, University of St. Andrews, St. Andrews, UK.