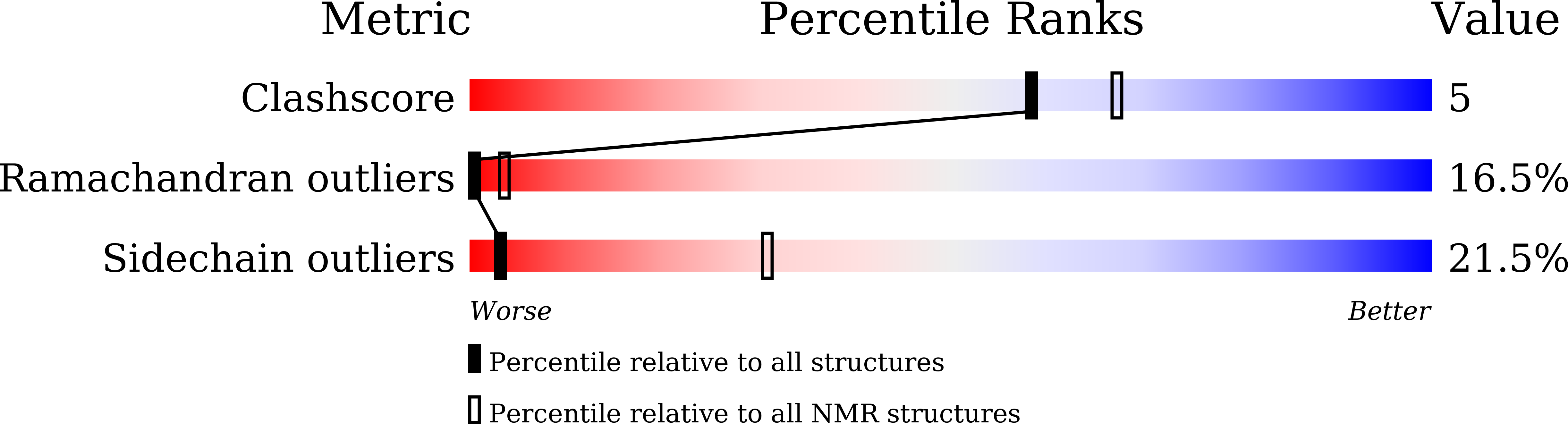The lantibiotic nukacin ISK-1 exists in an equilibrium between active and inactive lipid-II binding states.
Fujinami, D., Mahin, A.A., Elsayed, K.M., Islam, M.R., Nagao, J.I., Roy, U., Momin, S., Zendo, T., Kohda, D., Sonomoto, K.(2018) Commun Biol 1: 150-150
- PubMed: 30272026
- DOI: https://doi.org/10.1038/s42003-018-0150-3
- Primary Citation of Related Structures:
5Z5Q, 5Z5R - PubMed Abstract:
The lantibiotic nukacin ISK-1 exerts antimicrobial activity through binding to lipid II. Here, we perform NMR analyses of the structure of nukacin ISK-1 and the interaction with lipid II. Unexpectedly, nukacin ISK-1 exists in two structural states in aqueous solution, with an interconversion rate on a time scale of seconds. The two structures differ in the relative orientations of the two lanthionine rings, ring A and ring C. Chemical shift perturbation induced by the titration of lipid II reveals that only one state was capable of binding to lipid II. On the molecular surface of the active state, a multiple hydrogen-bonding site formed by amino acid residues in the ring A region is adjacent to a hydrophobic surface formed by residues in the ring C region, and we propose that these sites interact with the pyrophosphate moiety and the isoprene chain of the lipid II molecule, respectively.
Organizational Affiliation:
Division of Structural Biology, Medical Institute of Bioregulation, Kyushu University, Fukuoka, 812-8582, Japan.