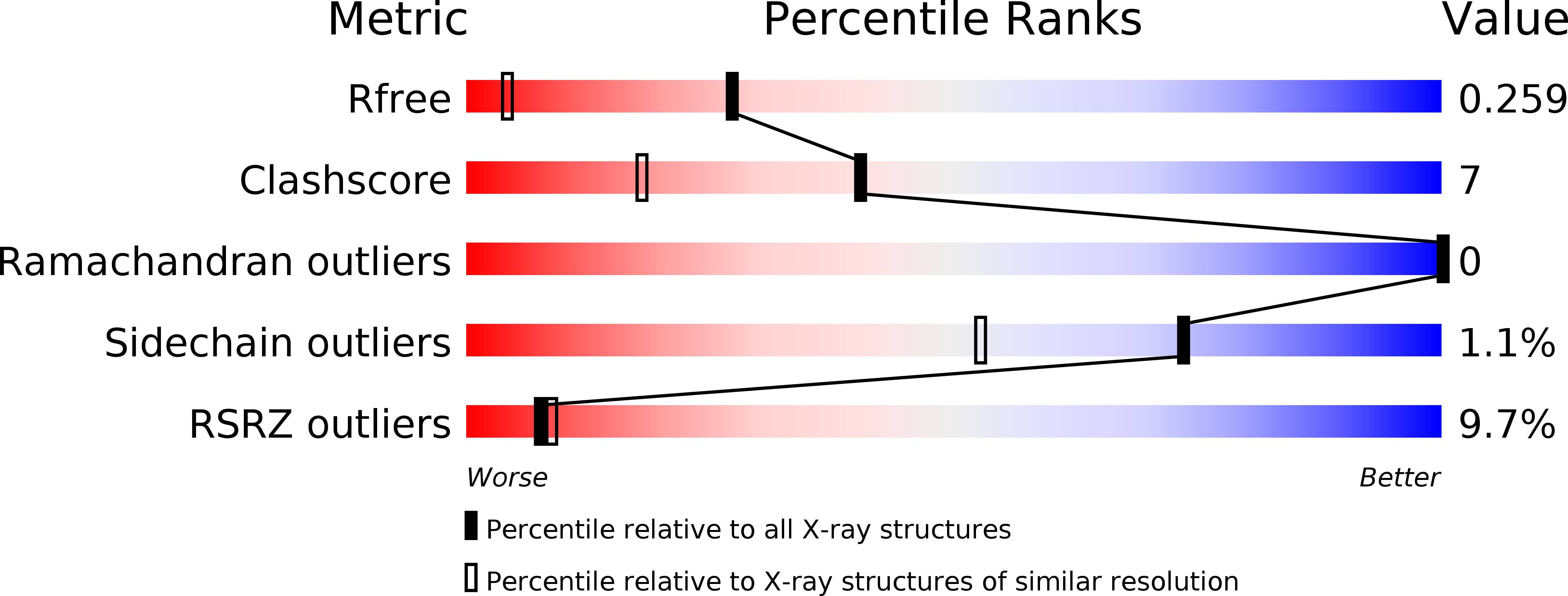
Dose and Schedule Determine Distinct Molecular Mechanisms Underlying the Efficacy of the p53-MDM2 Inhibitor HDM201.
Jeay, S., Ferretti, S., Holzer, P., Fuchs, J., Chapeau, E.A., Wartmann, M., Sterker, D., Romanet, V., Murakami, M., Kerr, G., Durand, E.Y., Gaulis, S., Cortes-Cros, M., Ruetz, S., Stachyra, T.M., Kallen, J., Furet, P., Wurthner, J., Guerreiro, N., Halilovic, E., Jullion, A., Kauffmann, A., Kuriakose, E., Wiesmann, M., Jensen, M.R., Hofmann, F., Sellers, W.R.(2018) Cancer Res 78: 6257-6267
- PubMed: 30135191
- DOI: https://doi.org/10.1158/0008-5472.CAN-18-0338
- Primary Citation of Related Structures:
5OC8 - PubMed Abstract:
Activation of p53 by inhibitors of the p53-MDM2 interaction is being pursued as a therapeutic strategy in p53 wild-type cancers. Here, we report distinct mechanisms by which the novel, potent, and selective inhibitor of the p53-MDM2 interaction HDM201 elicits therapeutic efficacy when applied at various doses and schedules. Continuous exposure of HDM201 led to induction of p21 and delayed accumulation of apoptotic cells. By comparison, high-dose pulses of HDM201 were associated with marked induction of PUMA and a rapid onset of apoptosis. shRNA screens identified PUMA as a mediator of the p53 response specifically in the pulsed regimen. Consistent with this, the single high-dose HDM201 regimen resulted in rapid and marked induction of PUMA expression and apoptosis together with downregulation of Bcl-xL in vivo Knockdown of Bcl-xL was identified as the top sensitizer to HDM201 in vitro , and Bcl-xL was enriched in relapsing tumors from mice treated with intermittent high doses of HDM201. These findings define a regimen-dependent mechanism by which disruption of MDM2-p53 elicits therapeutic efficacy when given with infrequent dosing. In an ongoing HDM201 trial, the observed exposure-response relationship indicates that the molecular mechanism elicited by pulse dosing is likely reproducible in patients. These data support the clinical comparison of daily and intermittent regimens of p53-MDM2 inhibitors. Significance: Pulsed high doses versus sustained low doses of the p53-MDM2 inhibitor HDM201 elicit a proapoptotic response from wild-type p53 cancer cells, offering guidance to current clinical trials with this and other drugs that exploit the activity of p53. Cancer Res; 78(21); 6257-67. ©2018 AACR .
Organizational Affiliation:
Disease Area Oncology, Novartis Institutes for BioMedical Research, Basel, Switzerland.