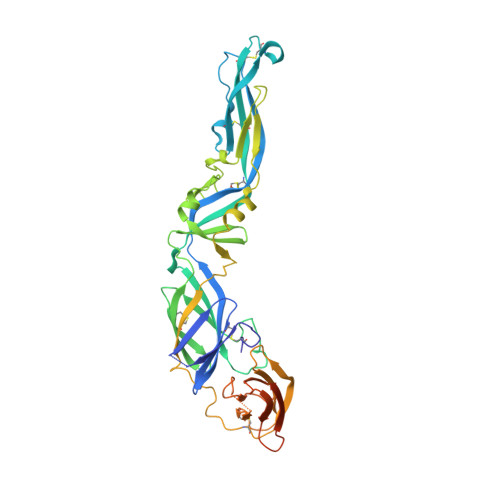
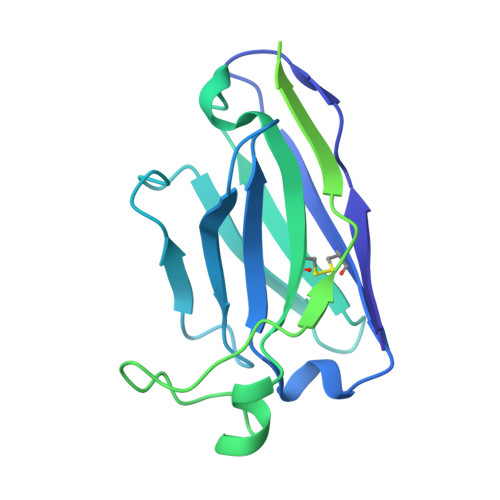
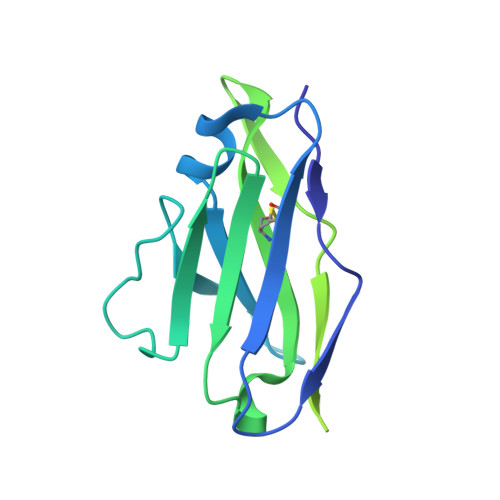
Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope.
Rouvinski, A., Dejnirattisai, W., Guardado-Calvo, P., Vaney, M.C., Sharma, A., Duquerroy, S., Supasa, P., Wongwiwat, W., Haouz, A., Barba-Spaeth, G., Mongkolsapaya, J., Rey, F.A., Screaton, G.R.(2017) Nat Commun 8: 15411-15411
- PubMed: 28534525
- DOI: https://doi.org/10.1038/ncomms15411
- Primary Citation of Related Structures:
5N09, 5N0A - PubMed Abstract:
A problem in the search for an efficient vaccine against dengue virus is the immunodominance of the fusion loop epitope (FLE), a segment of the envelope protein E that is buried at the interface of the E dimers coating mature viral particles. Anti-FLE antibodies are broadly cross-reactive but poorly neutralizing, displaying a strong infection enhancing potential. FLE exposure takes place via dynamic 'breathing' of E dimers at the virion surface. In contrast, antibodies targeting the E dimer epitope (EDE), readily exposed at the E dimer interface over the region of the conserved fusion loop, are very potent and broadly neutralizing. We here engineer E dimers locked by inter-subunit disulfide bonds, and show by X-ray crystallography and by binding to a panel of human antibodies that these engineered dimers do not expose the FLE, while retaining the EDE exposure. These locked dimers are strong immunogen candidates for a next-generation vaccine.
Organizational Affiliation:
Institut Pasteur, Département de Virologie, Unité de Virologie Structurale, 75724 Paris Cedex 15, France.