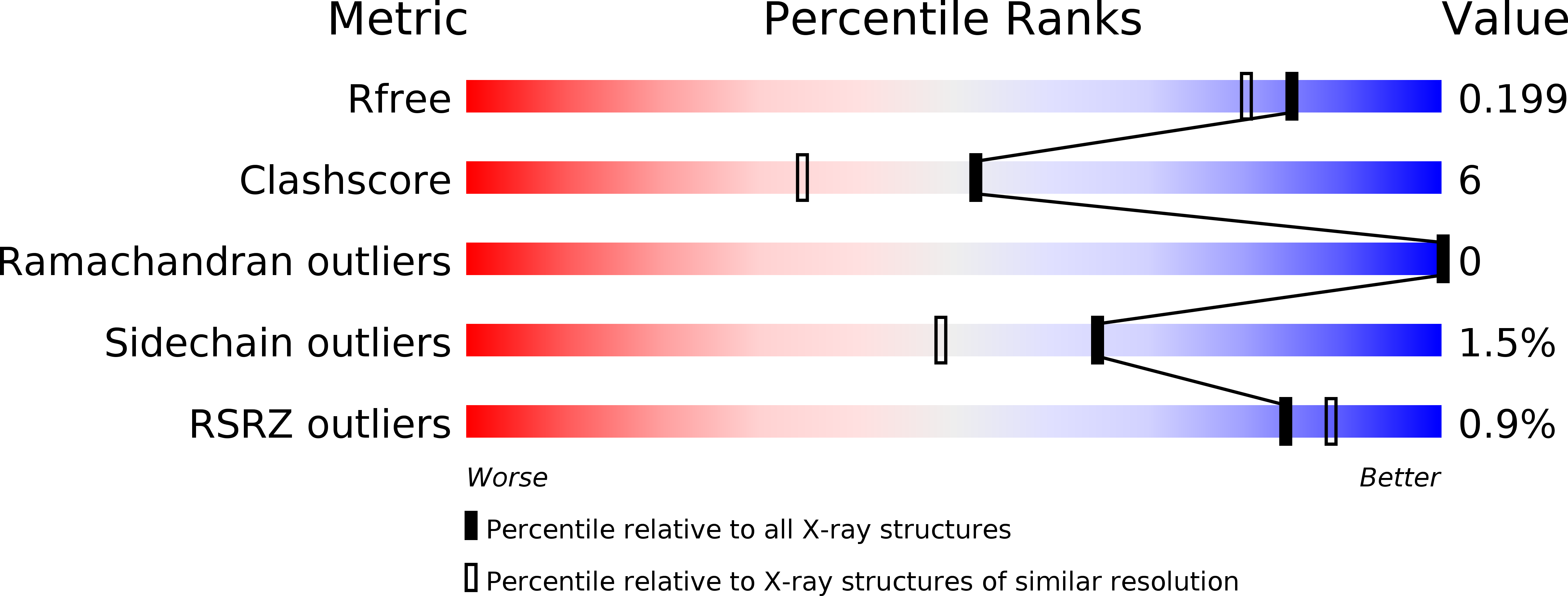
Structure and decoy-mediated inhibition of the SOX18/Prox1-DNA interaction.
Klaus, M., Prokoph, N., Girbig, M., Wang, X., Huang, Y.H., Srivastava, Y., Hou, L., Narasimhan, K., Kolatkar, P.R., Francois, M., Jauch, R.(2016) Nucleic Acids Res 44: 3922-3935
- PubMed: 26939885
- DOI: https://doi.org/10.1093/nar/gkw130
- Primary Citation of Related Structures:
4Y60 - PubMed Abstract:
The transcription factor (TF) SOX18 drives lymphatic vessel development in both embryogenesis and tumour-induced neo-lymphangiogenesis. Genetic disruption of Sox18 in a mouse model protects from tumour metastasis and established the SOX18 protein as a molecular target. Here, we report the crystal structure of the SOX18 DNA binding high-mobility group (HMG) box bound to a DNA element regulating Prox1 transcription. The crystals diffracted to 1.75Å presenting the highest resolution structure of a SOX/DNA complex presently available revealing water structure, structural adjustments at the DNA contact interface and non-canonical conformations of the DNA backbone. To explore alternatives to challenging small molecule approaches for targeting the DNA-binding activity of SOX18, we designed a set of five decoys based on modified Prox1-DNA. Four decoys potently inhibited DNA binding of SOX18 in vitro and did not interact with non-SOX TFs. Serum stability, nuclease resistance and thermal denaturation assays demonstrated that a decoy circularized with a hexaethylene glycol linker and terminal phosphorothioate modifications is most stable. This SOX decoy also interfered with the expression of a luciferase reporter under control of a SOX18-dependent VCAM1 promoter in COS7 cells. Collectively, we propose SOX decoys as potential strategy for inhibiting SOX18 activity to disrupt tumour-induced neo-lymphangiogenesis.
Organizational Affiliation:
Genome Regulation Laboratory, Drug Discovery Pipeline, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China Key Laboratory of Regenerative Biology, South China Institute for Stem Cell Biology and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, South China Institute for Stem Cell Biology and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China Institut für Chemie und Biochemie, Freie Universität Berlin, Thielallee 63, 14195 Berlin, Germany.