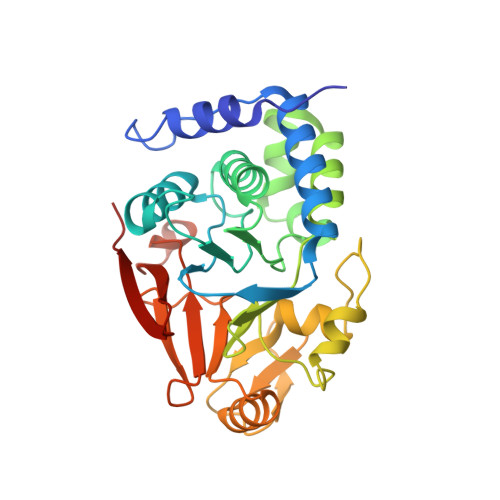
Structural and Functional Analysis of the GADD34:PP1 eIF2 alpha Phosphatase.
Choy, M.S., Yusoff, P., Lee, I.C., Newton, J.C., Goh, C.W., Page, R., Shenolikar, S., Peti, W.(2015) Cell Rep 11: 1885-1891
- PubMed: 26095357
- DOI: https://doi.org/10.1016/j.celrep.2015.05.043
- Primary Citation of Related Structures:
4XPN - PubMed Abstract:
The attenuation of protein synthesis via the phosphorylation of eIF2α is a major stress response of all eukaryotic cells. The growth-arrest- and DNA-damage-induced transcript 34 (GADD34) bound to the serine/threonine protein phosphatase 1 (PP1) is the necessary eIF2α phosphatase complex that returns mammalian cells to normal protein synthesis following stress. The molecular basis by which GADD34 recruits PP1 and its substrate eIF2α are not fully understood, hindering our understanding of the remarkable selectivity of the GADD34:PP1 phosphatase for eIF2α. Here, we report detailed structural and functional analyses of the GADD34:PP1 holoenzyme and its recruitment of eIF2α. The data highlight independent interactions of PP1 and eIF2α with GADD34, demonstrating that GADD34 functions as a scaffold both in vitro and in cells. This work greatly enhances our molecular understanding of a major cellular eIF2α phosphatase and establishes the foundation for future translational work.
Organizational Affiliation:
Department of Molecular Pharmacology, Physiology and Biotechnology, Brown University, Providence, RI 02912, USA.