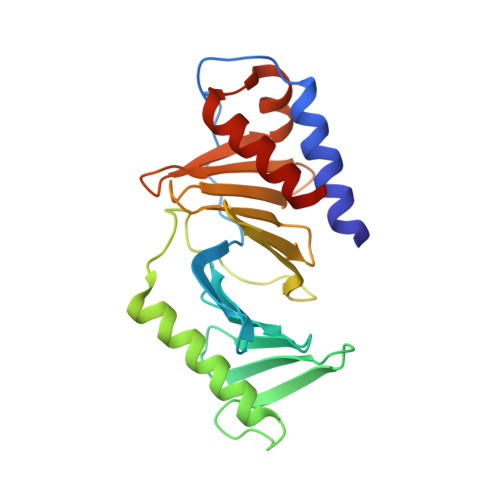
Structural analysis of the polo-box domain of human Polo-like kinase 2
Kim, J.H., Ku, B., Lee, K.S., Kim, S.J.(2015) Proteins 83: 1201-1208
- PubMed: 25846005
- DOI: https://doi.org/10.1002/prot.24804
- Primary Citation of Related Structures:
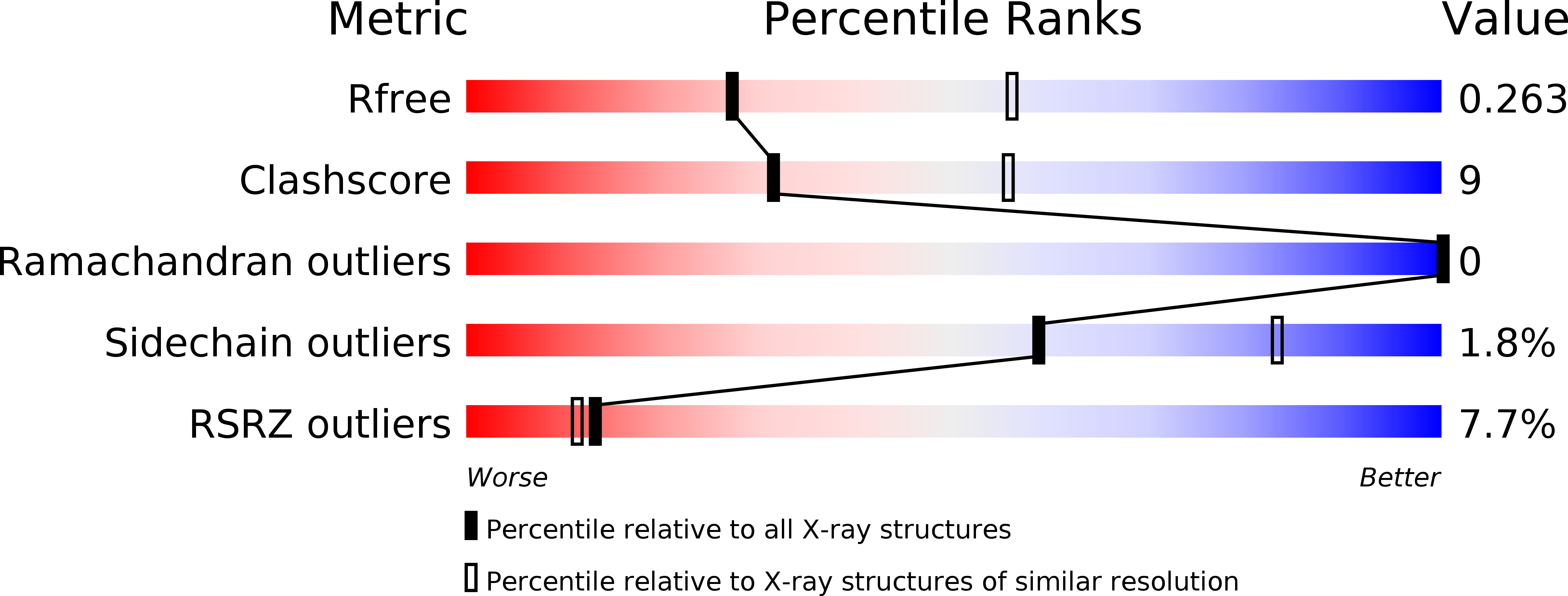
4XB0 - PubMed Abstract:
Polo-like kinases (Plks) are the key regulators of cell cycle progression, the members of which share a kinase domain and a polo-box domain (PBD) that serves as a protein-binding module. While Plk1 is a promising target for antitumor therapy, Plk2 is regarded as a tumor suppressor even though the two Plks commonly recognize the S-pS/T-P motif through their PBD. Herein, we report the crystal structure of the PBD of Plk2 at 2.7 Å. Despite the overall structural similarity with that of Plk1 reflecting their high sequence homology, the crystal structure also contains its own features including the highly ordered loop connecting two subdomains and the absence of 310 -helices in the N-terminal region unlike the PBD of Plk1. Based on the three-dimensional structure, we furthermore could model its interaction with two types of phosphopeptides, one of which was previously screened as the optimal peptide for the PBD of Plk2.
Organizational Affiliation:
Functional Genomics Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, 305-806, Korea.