Crystal structure of Methylobacterium extorquens malate dehydrogenase complexed with oxaloacetate and adenosine-5-diphosphoribose
Gonzalez, J.M., Marti-Arbona, R., Unkefer, C.J.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Malate dehydrogenase | 320 | Methylorubrum extorquens PA1 | Mutation(s): 0 Gene Names: mdh, Mext_1643 EC: 1.1.1.37 | 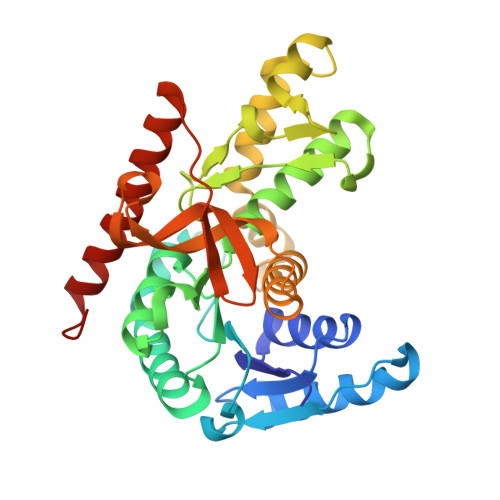 | |
UniProt | |||||
Find proteins for A9W386 (Methylorubrum extorquens (strain PA1)) Explore A9W386 Go to UniProtKB: A9W386 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A9W386 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| APR Query on APR | E [auth A] | ADENOSINE-5-DIPHOSPHORIBOSE C15 H23 N5 O14 P2 SRNWOUGRCWSEMX-KEOHHSTQSA-N |  | ||
| OAA Query on OAA | D [auth A] | OXALOACETATE ION C4 H3 O5 KHPXUQMNIQBQEV-UHFFFAOYSA-M |  | ||
| CA Query on CA | B [auth A], C [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 108.396 | α = 90 |
| b = 108.396 | β = 90 |
| c = 104.014 | γ = 120 |
| Software Name | Purpose |
|---|---|
| ADSC | data collection |
| PHASER | phasing |
| REFMAC | refinement |
| XDS | data reduction |
| SCALA | data scaling |