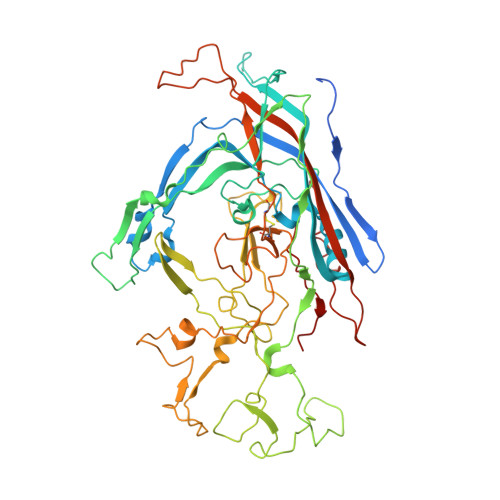
Global Displacement of Canine Parvovirus by a Host-Adapted Variant: Structural Comparison between Pandemic Viruses with Distinct Host Ranges.
Organtini, L.J., Allison, A.B., Lukk, T., Parrish, C.R., Hafenstein, S.(2015) J Virol 89: 1909-1912
- PubMed: 25410876
- DOI: https://doi.org/10.1128/JVI.02611-14
- Primary Citation of Related Structures:
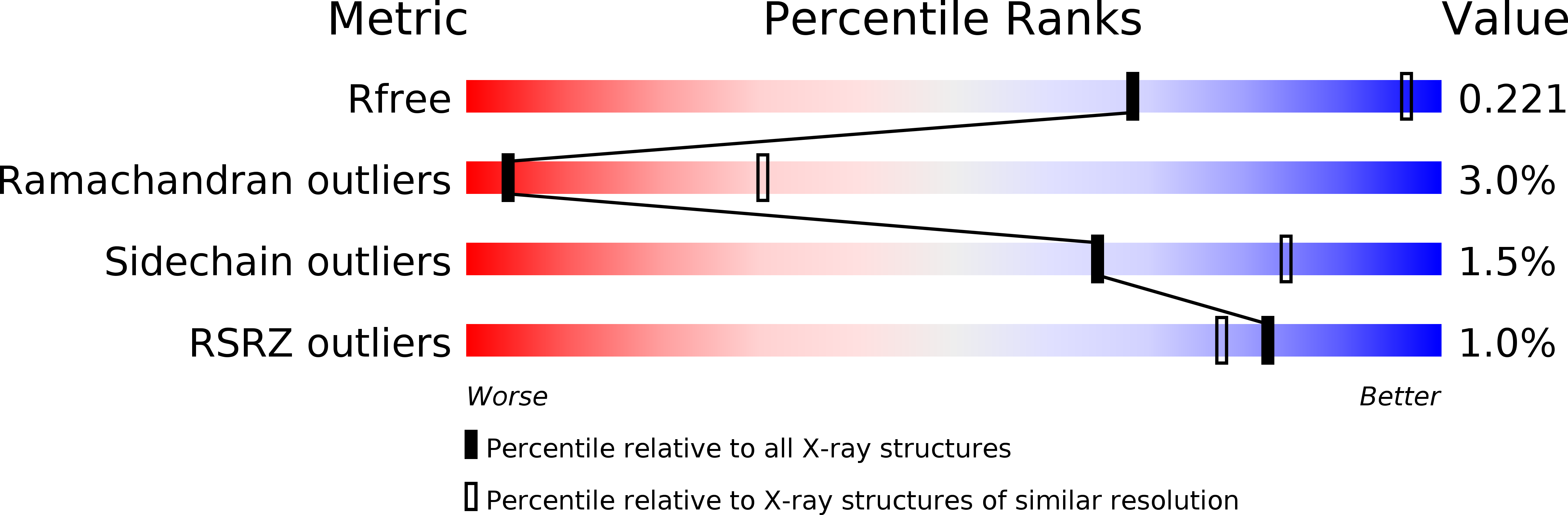
4QYK - PubMed Abstract:
Canine parvovirus type 2 (CPV-2) emerged in 1978 and spread worldwide within 2 years. Subsequently, CPV-2 was completely replaced by the variant CPV-2a, which is characterized by four specific capsid (VP2) mutations. The X-ray crystal structure of the CPV-2a capsid shows that each mutation confers small local changes. The loss of a hydrogen bond and introduction of a glycine residue likely introduce flexibility to sites that control interactions with the host receptor, antibodies, and sialic acids.
Organizational Affiliation:
Department of Medicine, The Pennsylvania State University College of Medicine, Hershey, Pennsylvania, USA.