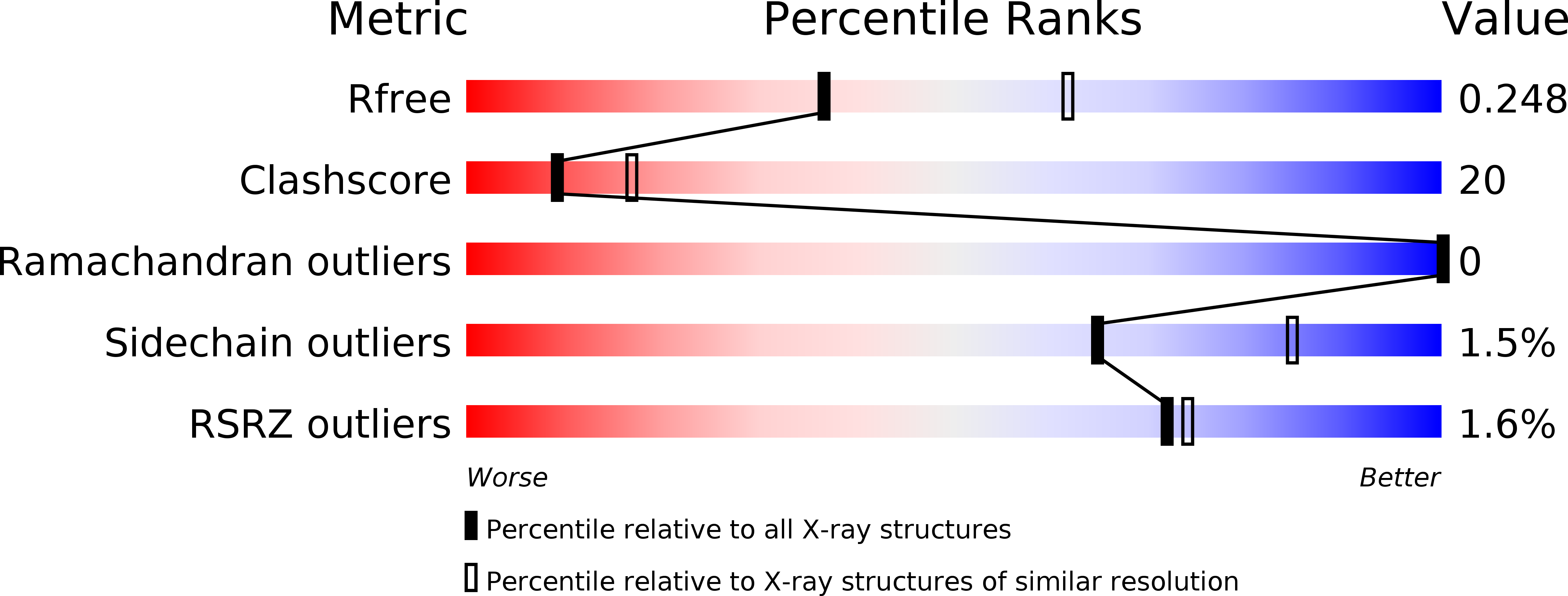
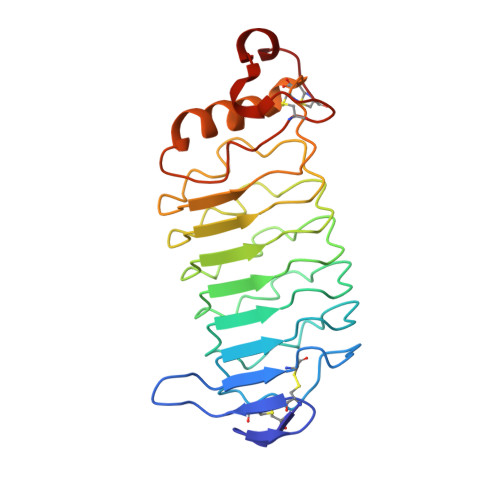
Selection of the lamprey VLRC antigen receptor repertoire.
Holland, S.J., Gao, M., Hirano, M., Iyer, L.M., Luo, M., Schorpp, M., Cooper, M.D., Aravind, L., Mariuzza, R.A., Boehm, T.(2014) Proc Natl Acad Sci U S A 111: 14834-14839
- PubMed: 25228760
- DOI: https://doi.org/10.1073/pnas.1415655111
- Primary Citation of Related Structures:
4PO4 - PubMed Abstract:
The alternative adaptive immune system of jawless vertebrates is based on different isotypes of variable lymphocyte receptors (VLRs) that are composed of leucine-rich repeats (LRRs) and expressed by distinct B- and T-like lymphocyte lineages. VLRB is expressed by B-like cells, whereas VLRA and VLRC are expressed by two T-like lineages that develop in the thymoid, a thymus-like structure in lamprey larvae. In each case, stepwise combinatorial insertions of different types of short donor LRR cassettes into incomplete germ-line genes are required to generate functional VLR gene assemblies. It is unknown, however, whether the diverse repertoires of VLRs that are expressed by peripheral blood lymphocytes are shaped by selection after their assembly. Here, we identify signatures of selection in the peripheral repertoire of VLRC antigen receptors that are clonally expressed by one of the T-like cell types in lampreys. Selection strongly favors VLRC molecules containing four internal variable leucine-rich repeat (LRRV) modules, although VLRC assemblies encoding five internal modules are initially equally frequent. In addition to the length selection, VLRC molecules in VLRC(+) peripheral lymphocytes exhibit a distinct pattern of high entropy sites in the N-terminal LRR1 module, which is inserted next to the germ-line-encoded LRRNT module. This is evident in comparisons to VLRC gene assemblies found in the thymoid and to VLRC gene assemblies found in some VLRA(+) cells. Our findings are the first indication to our knowledge that selection operates on a VLR repertoire and provide a framework to establish the mechanism by which this selection occurs during development of the VLRC(+) lymphocyte lineage.
Organizational Affiliation:
Department of Developmental Immunology, Max Planck Institute of Immunobiology and Epigenetics, 79108 Freiburg, Germany;