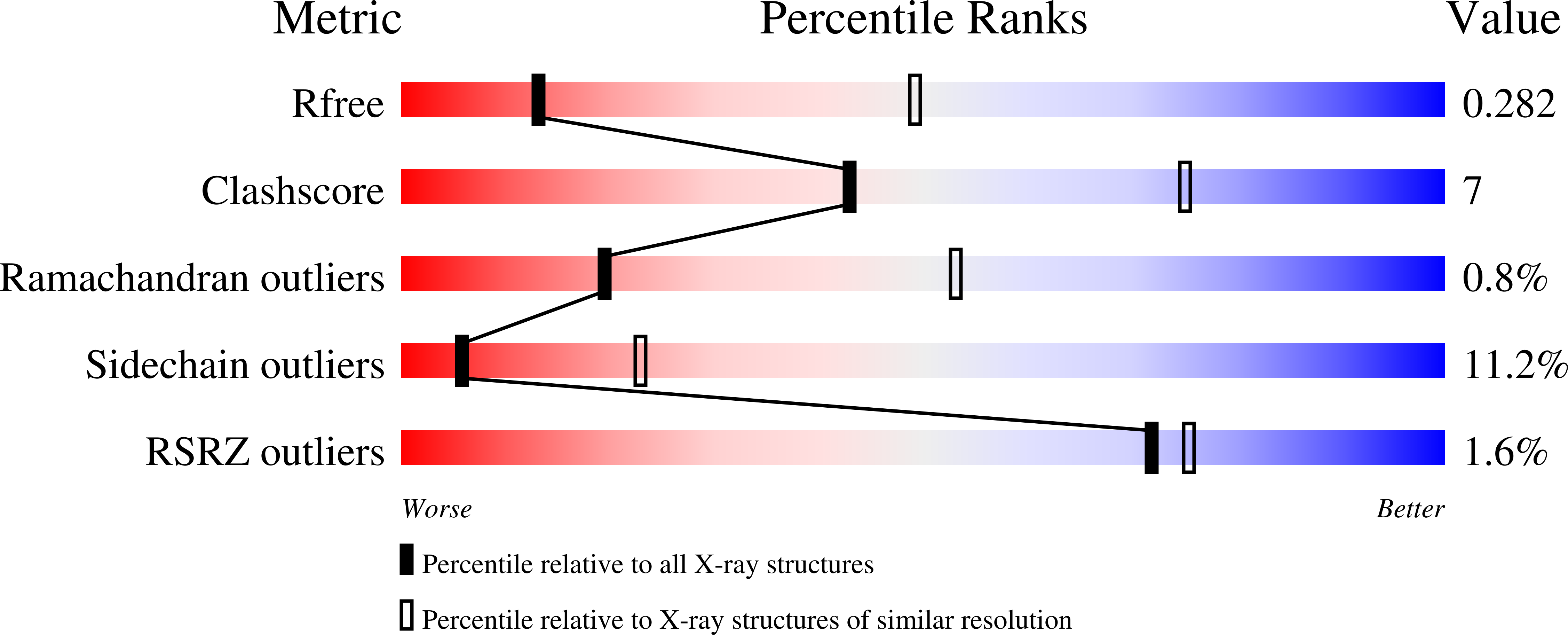
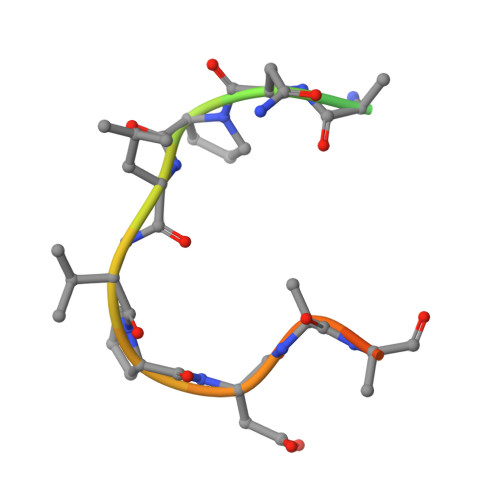
Structure of a C. perfringens Enterotoxin Mutant in Complex with a Modified Claudin-2 Extracellular Loop 2.
Yelland, T.S., Naylor, C.E., Bagoban, T., Savva, C.G., Moss, D.S., McClane, B.A., Blasig, I.E., Popoff, M., Basak, A.K.(2014) J Mol Biol 426: 3134-3147
- PubMed: 25020226
- DOI: https://doi.org/10.1016/j.jmb.2014.07.001
- Primary Citation of Related Structures:
3ZIW, 3ZIX, 4P5H - PubMed Abstract:
CPE (Clostridium perfringens enterotoxin) is the major virulence determinant for C. perfringens type-A food poisoning, the second most common bacterial food-borne illness in the UK and USA. After binding to its receptors, which include particular human claudins, the toxin forms pores in the cell membrane. The mature pore apparently contains a hexamer of CPE, claudin and, possibly, occludin. The combination of high binding specificity with cytotoxicity has resulted in CPE being investigated, with some success, as a targeted cytotoxic agent for oncotherapy. In this paper, we present the X-ray crystallographic structure of CPE in complex with a peptide derived from extracellular loop 2 of a modified, CPE-binding Claudin-2, together with high-resolution native and pore-formation mutant structures. Our structure provides the first atomic-resolution data on any part of a claudin molecule and reveals that claudin's CPE-binding fingerprint (NPLVP) is in a tight turn conformation and binds, as expected, in CPE's C-terminal claudin-binding groove. The leucine and valine residues insert into the binding groove while the first residue, asparagine, tethers the peptide via an interaction with CPE's aspartate 225 and the two prolines are required to maintain the tight turn conformation. Understanding the structural basis of the contribution these residues make to binding will aid in engineering CPE to target tumor cells.
Organizational Affiliation:
Department of Biological Sciences, Birkbeck College, London WC1E 7HX, UK.