Structural and kinetic properties of Rhodobacter sphaeroides photosynthetic reaction centers containing exclusively Zn-coordinated bacteriochlorophyll as bacteriochlorin cofactors.
Saer, R.G., Pan, J., Hardjasa, A., Lin, S., Rosell, F., Mauk, A.G., Woodbury, N.W., Murphy, M.E., Beatty, J.T.(2014) Biochim Biophys Acta 1837: 366-374
- PubMed: 24316146
- DOI: https://doi.org/10.1016/j.bbabio.2013.11.015
- Primary Citation of Related Structures:
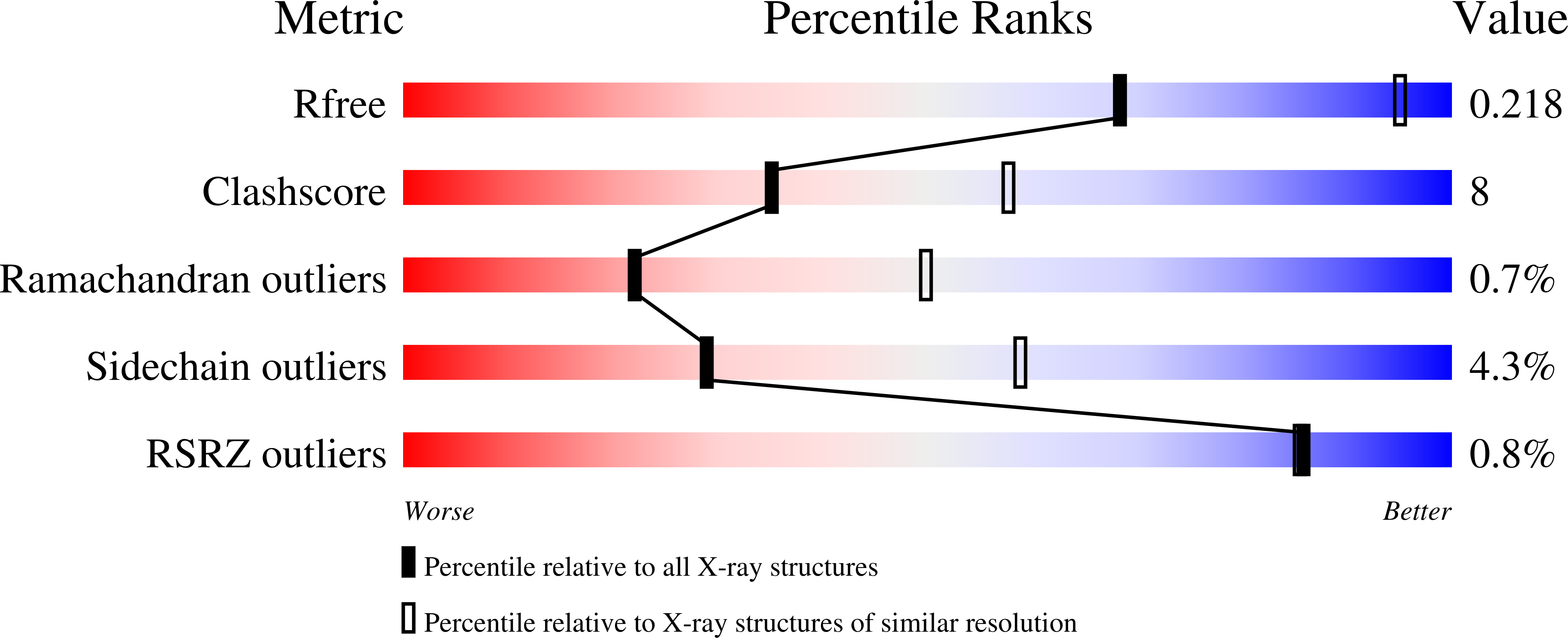
4N7K, 4N7L - PubMed Abstract:
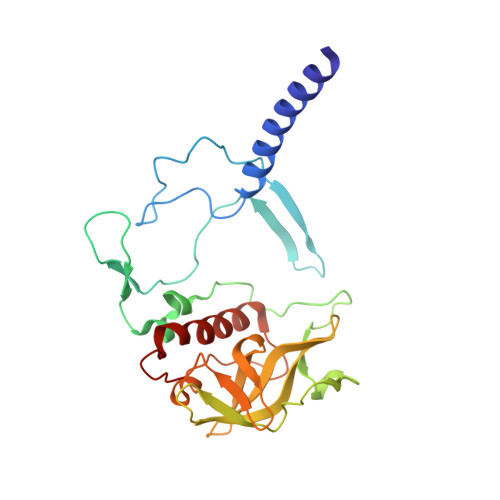
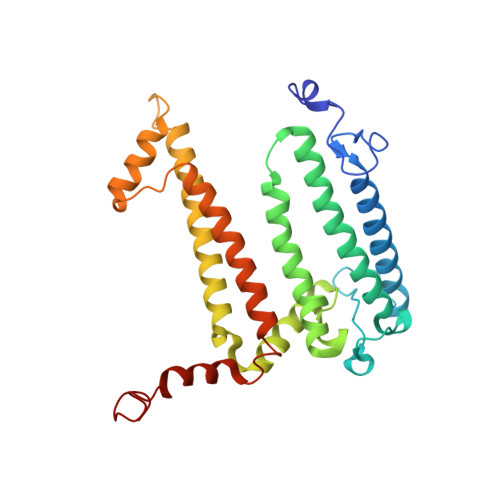
The Zn-BChl-containing reaction center (RC) produced in a bchD (magnesium chelatase) mutant of Rhodobacter sphaeroides assembles with six Zn-bacteriochlorophylls (Zn-BChls) in place of four Mg-containing bacteriochlorophylls (BChls) and two bacteriopheophytins (BPhes). This protein presents unique opportunities for studying biological electron transfer, as Zn-containing chlorins can exist in 4-, 5-, and (theoretically) 6-coordinate states within the RC. In this paper, the electron transfer perturbations attributed exclusively to coordination state effects are separated from those attributed to the presence, absence, or type of metal in the bacteriochlorin at the HA pocket of the RC. The presence of a 4-coordinate Zn(2+) ion in the HA bacteriochlorin instead of BPhe results in a small decrease in the rates of the P*→P(+)HA(-)→P(+)QA(-) electron transfer, and the charge separation yield is not greatly perturbed; however coordination of the Zn(2+) by a fifth ligand provided by a histidine residue results in a larger rate decrease and yield loss. We also report the first crystal structure of a Zn-BChl-containing RC, confirming that the HA Zn-BChl was either 4- or 5-coordinate in the two types of Zn-BChl-containing RCs studied here. Interestingly, a large degree of disorder, in combination with a relatively weak anomalous difference electron density was found in the HB pocket. These data, in combination with spectroscopic results, indicate partial occupancy of this binding pocket. These findings provide insights into the use of BPhe as the bacteriochlorin pigment of choice at HA in both BChl- and Zn-BChl-containing RCs found in nature.
Organizational Affiliation:
Department of Microbiology and Immunology, The University of British Columbia, 2350 Health Sciences Mall, Vancouver, BC V6T 1Z3, Canada.