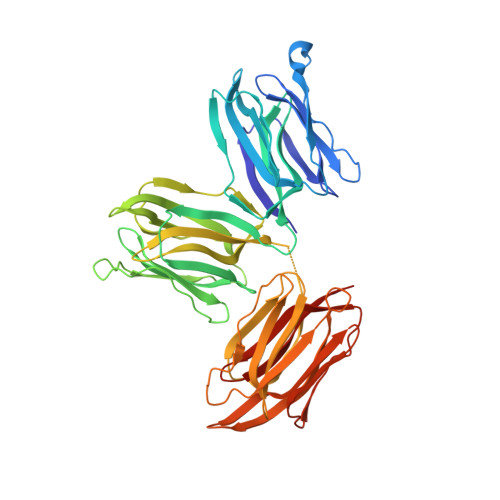
Crystal structure of Parkia biglobosa seed lectin (PBL) in complex with methyl alpha D-mannopyranoside
Teixeira, C.S., Rocha, B.A.M., Silva, H.C., Bari, A.U., Barroso-Neto, I.T., Santiago, M.Q., Nagano, C.S., Nascimento, K.S., Debray, H., Delatorre, P., Cavada, B.S.To be published.