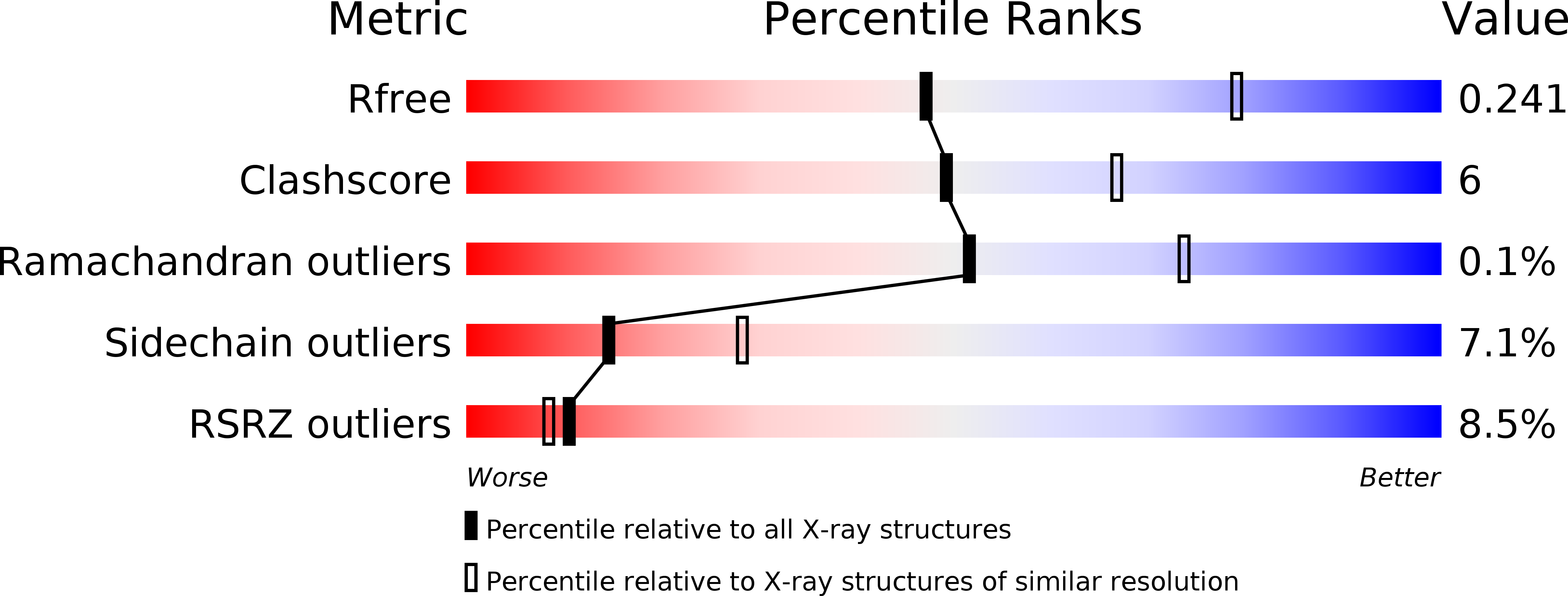
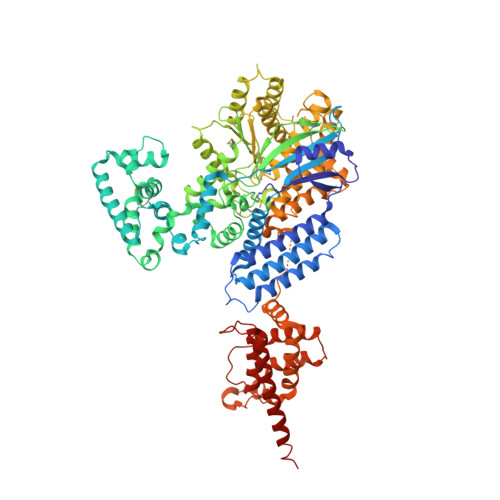
Identification and structural characterization of a legionella phosphoinositide phosphatase.
Toulabi, L., Wu, X., Cheng, Y., Mao, Y.(2013) J Biol Chem 288: 24518-24527
- PubMed: 23843460
- DOI: https://doi.org/10.1074/jbc.M113.474239
- Primary Citation of Related Structures:
4JZA - PubMed Abstract:
Bacterial pathogen Legionella pneumophila is the causative agent of Legionnaires' disease, which is associated with intracellular replication of the bacteria in macrophages of human innate immune system. Recent studies indicate that pathogenic bacteria can subvert host cell phosphoinositide (PI) metabolism by translocated virulence effectors. However, in which manner Legionella actively exploits PI lipids to benefit its infection is not well characterized. Here we report that L. pneumophila encodes an effector protein, named SidP, that functions as a PI-3-phosphatase specifically hydrolyzing PI(3)P and PI(3,5)P2 in vitro. This activity of SidP rescues the growth phenotype of a yeast strain defective in PI(3)P phosphatase activity. Crystal structure of SidP orthologue from Legionella longbeachae reveals that this unique PI-3-phosphatase is composed of three distinct domains: a large catalytic domain, an appendage domain that is inserted into the N-terminal portion of the catalytic domain, and a C-terminal α-helical domain. SidP has a small catalytic pocket that presumably provides substrate specificity by limiting the accessibility of bulky PIs with multiple phosphate groups. Together, our identification of a unique family of Legionella PI phosphatases highlights a common scheme of exploiting host PI lipids in many intracellular bacterial pathogen infections.
Organizational Affiliation:
Weill Institute for Cell and Molecular Biology and Department of Molecular Biology and Genetics, Cornell University, Ithaca, New York 14853, USA.