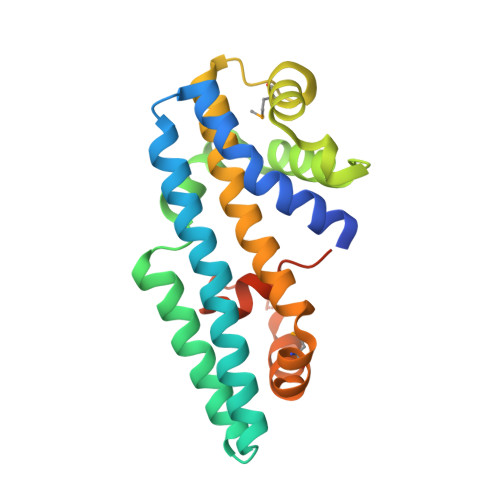
Structure of the Helicobacter pylori CagA oncoprotein bound to the human tumor suppressor ASPP2.
Nesic, D., Buti, L., Lu, X., Stebbins, C.E.(2014) Proc Natl Acad Sci U S A 111: 1562-1567
- PubMed: 24474782
- DOI: https://doi.org/10.1073/pnas.1320631111
- Primary Citation of Related Structures:
4IRV - PubMed Abstract:
The Cytotoxin associated gene A (CagA) protein of Helicobacter pylori is associated with increased virulence and risk of cancer. Recent proteomic studies have demonstrated an association of CagA with the human tumor suppressor Apoptosis-stimulating Protein of p53-2 (ASPP2). We present here a genetic, biochemical, and structural analysis of CagA with ASPP2. Domain delineation of the 120-kDa CagA protein revealed a stable N-terminal subdomain that was used in a yeast two-hybrid screen that identified the proline-rich domain of ASPP2 as a host cellular target. Biochemical experiments confirm this interaction. The cocrystal structure to 2.0-Å resolution of this N-terminal subdomain of CagA with a 7-kDa proline-rich sequence of ASPP2 reveals that this domain of CagA forms a highly specialized three-helix bundle, with large insertions in the loops connecting the helices. These insertions come together to form a deep binding cleft for a highly conserved 20-aa peptide of ASPP2. ASPP2 forms an extended helix in this groove of CagA, burying more than 1,000 Å(2) of surface area. This interaction is disrupted in vitro and in vivo by structure-based, loss-of-contact point mutations of key residues in either CagA or ASPP2. Disruption of CagA and ASPP2 binding alters the function of ASPP2 and leads to the decreased survival of H. pylori-infected cells.
Organizational Affiliation:
Laboratory of Structural Microbiology, The Rockefeller University, New York, NY 10065.