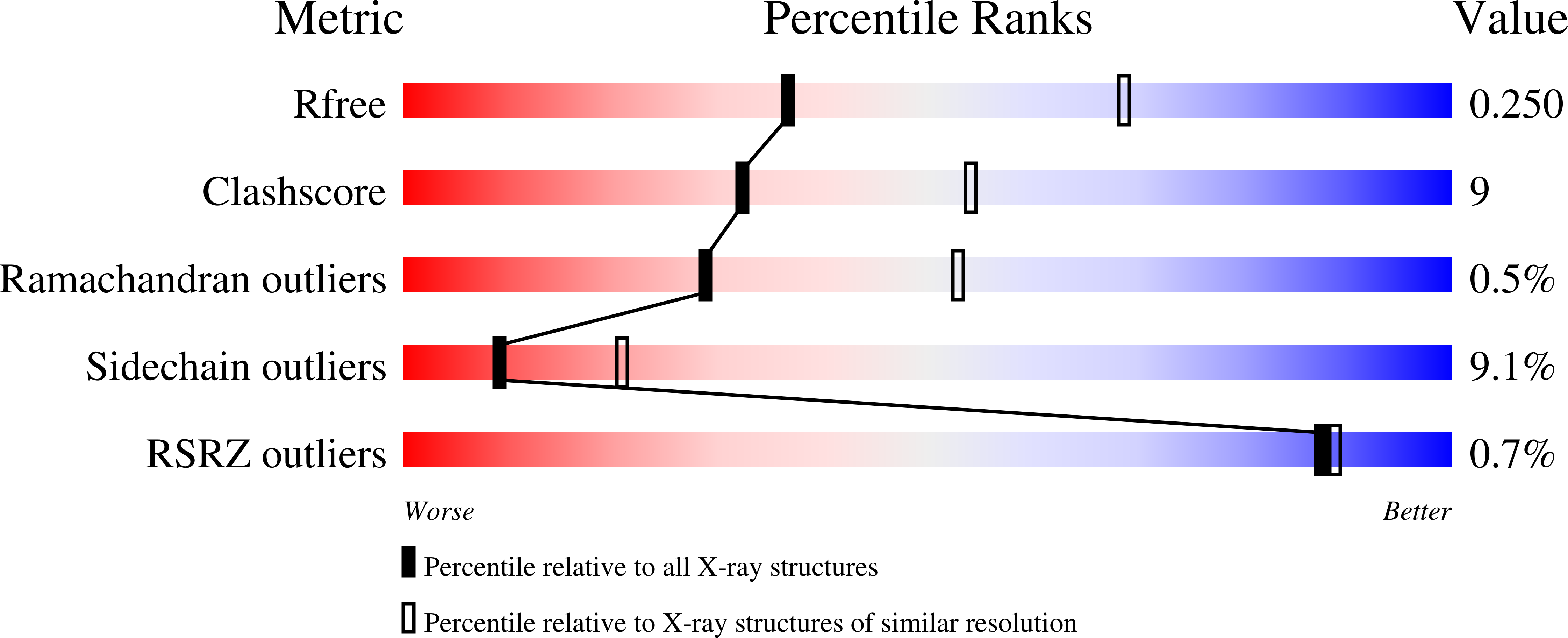
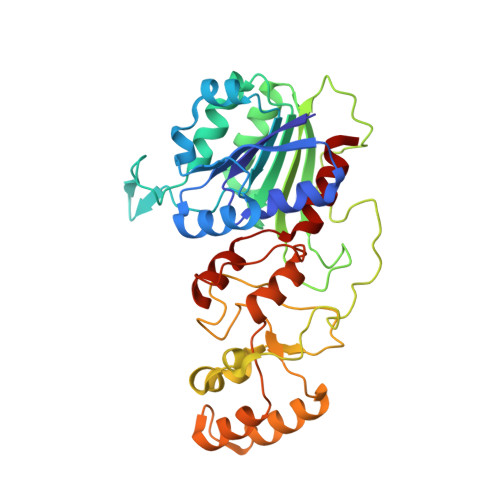
Functional and structural characterization of DNMT2 from Spodoptera frugiperda.
Li, S., Du, J., Yang, H., Yin, J., Ding, J., Zhong, J.(2013) J Mol Cell Biol 5: 64-66
- PubMed: 23103599
- DOI: https://doi.org/10.1093/jmcb/mjs057
- Primary Citation of Related Structures:
4H0N