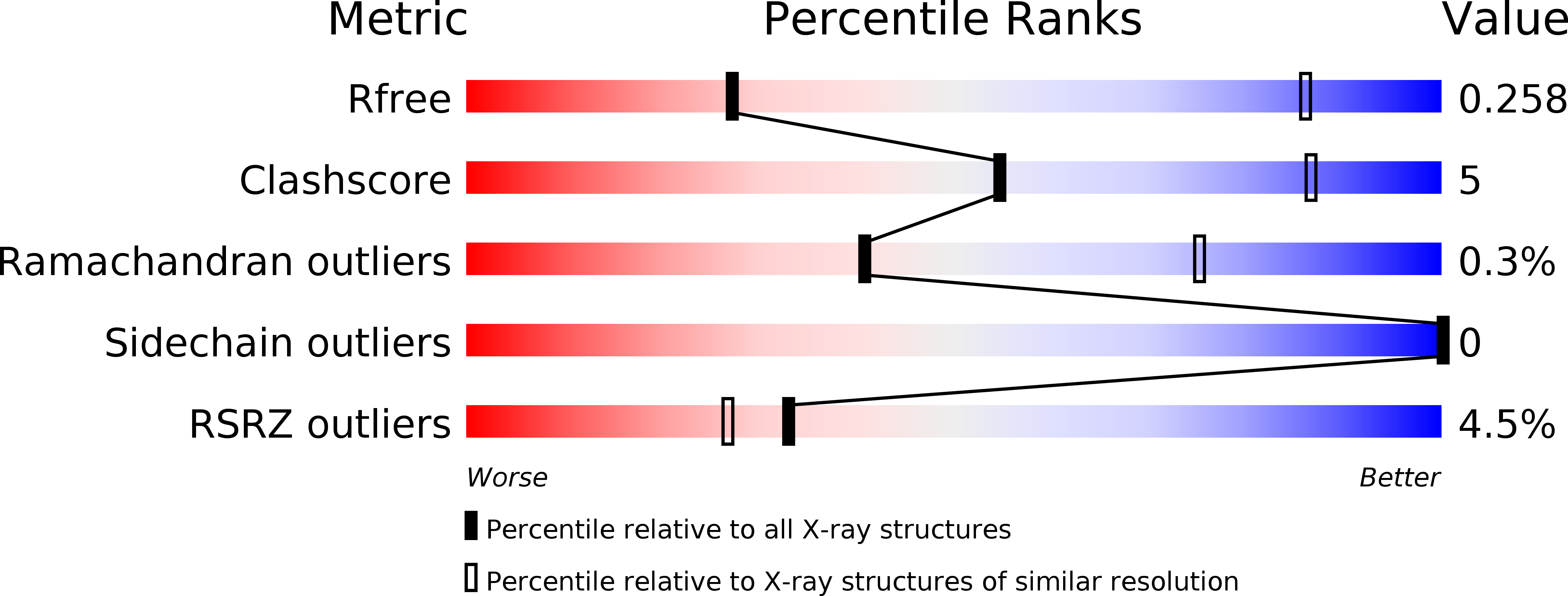
Structure of the st. Louis encephalitis virus postfusion envelope trimer.
Luca, V.C., Nelson, C.A., Fremont, D.H.(2013) J Virol 87: 818-828
- PubMed: 23115296
- DOI: https://doi.org/10.1128/JVI.01950-12
- Primary Citation of Related Structures:
4FG0 - PubMed Abstract:
St. Louis encephalitis virus (SLEV) is a mosquito-borne flavivirus responsible for several human encephalitis outbreaks over the last 80 years. Mature flavivirus virions are coated with dimeric envelope (E) proteins that mediate attachment and fusion with host cells. E is a class II fusion protein, the hallmark of which is a distinct dimer-to-trimer rearrangement that occurs upon endosomal acidification and insertion of hydrophobic fusion peptides into the endosomal membrane. Herein, we report the crystal structure of SLEV E in the posfusion trimer conformation. The structure revealed specific features that differentiate SLEV E from trimers of related flavi- and alphaviruses. SLEV E fusion loops have distinct intermediate spacing such that they are positioned further apart than previously observed in flaviviruses but closer together than Semliki Forest virus, an alphavirus. Domains II and III (DII and DIII) of SLEV E also adopt different angles relative to DI, which suggests that the DI-DII joint may accommodate spheroidal motions. However, trimer interfaces are well conserved among flaviviruses, so it is likely the differences observed represent structural features specific to SLEV function. Analysis of surface potentials revealed a basic platform underneath flavivirus fusion loops that may interact with the anionic lipid head groups found in membranes. Taken together, these results highlight variations in E structure and assembly that may direct virus-specific interactions with host determinants to influence pathogenesis.
Organizational Affiliation:
Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO, USA.