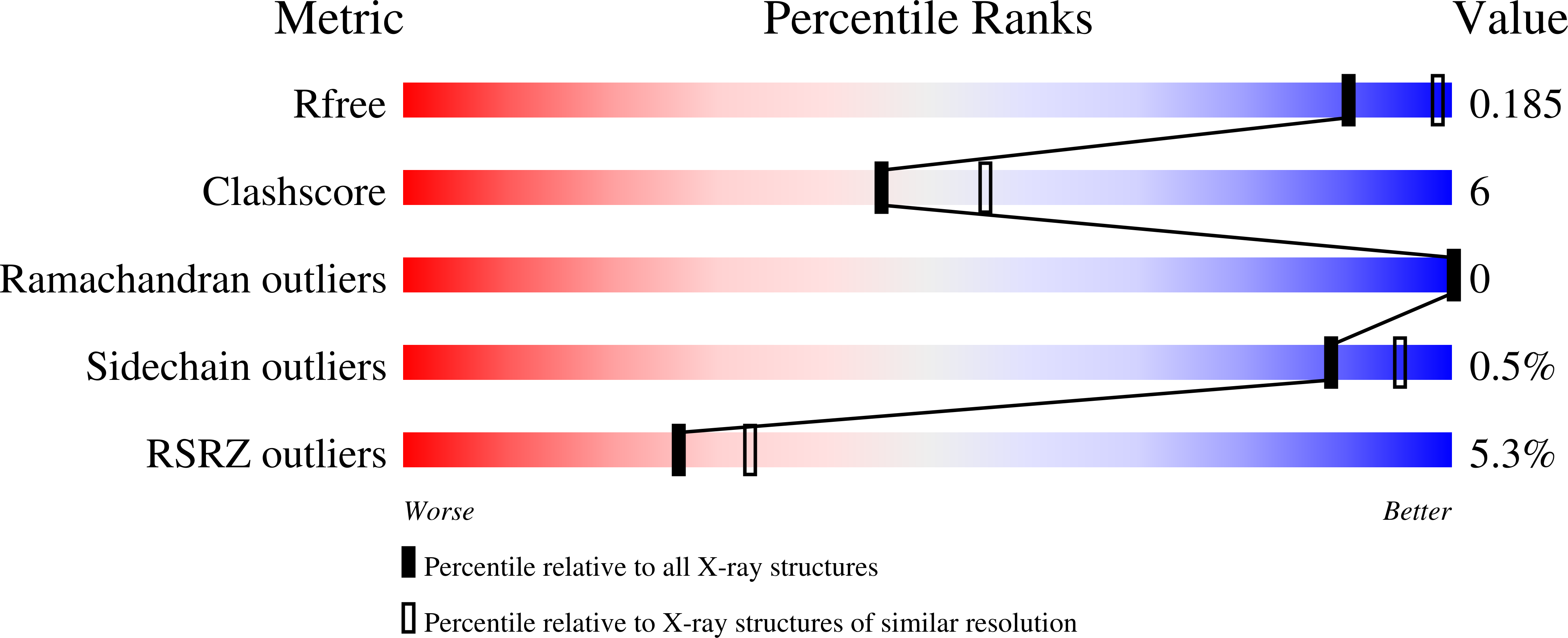
Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme glucose-1-phosphate thymidylyltransferase (RfbA).
Baumgartner, J., Lee, J., Halavaty, A.S., Minasov, G., Anderson, W.F., Kuhn, M.L.(2017) Acta Crystallogr F Struct Biol Commun 73: 621-628
- PubMed: 29095156
- DOI: https://doi.org/10.1107/S2053230X17015357
- Primary Citation of Related Structures:
4ECM - PubMed Abstract:
L-Rhamnose is a ubiquitous bacterial cell-wall component. The biosynthetic pathway for its precursor dTDP-L-rhamnose is not present in humans, which makes the enzymes of the pathway potential drug targets. In this study, the three-dimensional structure of the first protein of this pathway, glucose-1-phosphate thymidylyltransferase (RfbA), from Bacillus anthracis was determined. In other organisms this enzyme is referred to as RmlA. RfbA was co-crystallized with the products of the enzymatic reaction, dTDP-α-D-glucose and pyrophosphate, and its structure was determined at 2.3 Å resolution. This is the first reported thymidylyltransferase structure from a Gram-positive bacterium. RfbA shares overall structural characteristics with known RmlA homologs. However, RfbA exhibits a shorter sequence at its C-terminus, which results in the absence of three α-helices involved in allosteric site formation. Consequently, RfbA was observed to exhibit a quaternary structure that is unique among currently reported glucose-1-phosphate thymidylyltransferase bacterial homologs. These structural analyses suggest that RfbA may not be allosterically regulated in some organisms and is structurally distinct from other RmlA homologs.
Organizational Affiliation:
Department of Chemistry and Biochemistry, San Francisco State University, USA.