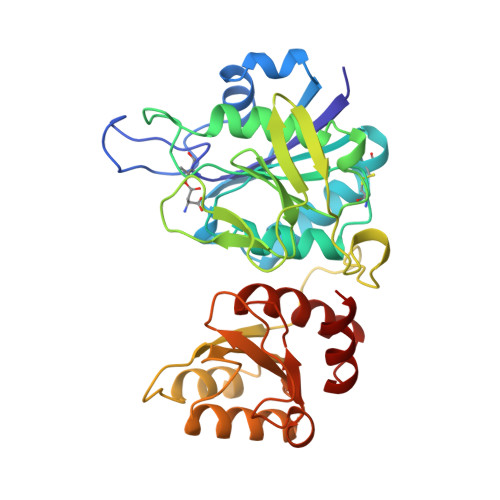
A covalently bound catalytic intermediate in Escherichia coli asparaginase: crystal structure of a Thr-89-Val mutant.
Palm, G.J., Lubkowski, J., Derst, C., Schleper, S., Rohm, K.H., Wlodawer, A.(1996) FEBS Lett 390: 211-216
- PubMed: 8706862
- DOI: https://doi.org/10.1016/0014-5793(96)00660-6
- Primary Citation of Related Structures:
4ECA - PubMed Abstract:
Escherichia coli asparaginase II catalyzes the hydrolysis of L-asparagine to L-aspartate via a threonine-bound acyl-enzyme intermediate. A nearly inactive mutant in which one of the active site threonines, Thr-89, was replaced by valine was constructed, expressed, and crystallized. Its structure, solved at 2.2 A resolution, shows high overall similarity to the wild-type enzyme, but an aspartyl moiety is covalently bound to Thr-12, resembling a reaction intermediate. Kinetic analysis confirms the deacylation deficiency, which is also explained on a structural basis. The previously identified oxyanion hole is described in more detail.
Organizational Affiliation:
Macromolecular Structure Laboratory, NCI-Frederick Cancer Research and Development Center, Frederick, MD 21702-1201, USA.