The X-Ray Structure of Nccx from Cupriavidus Metallidurans 31A Illustrates Potential Dangers of Detergent Solubilization When Generating and Interpreting Crystal Structures of Membrane Proteins.
Ziani, W., Maillard, A.P., Petit-Hartlein, I., Garnier, N., Crouzy, S., Girard, E., Coves, J.(2014) J Biol Chem 289: 31160
- PubMed: 25258316
- DOI: https://doi.org/10.1074/jbc.M114.586537
- Primary Citation of Related Structures:
4CLV - PubMed Abstract:
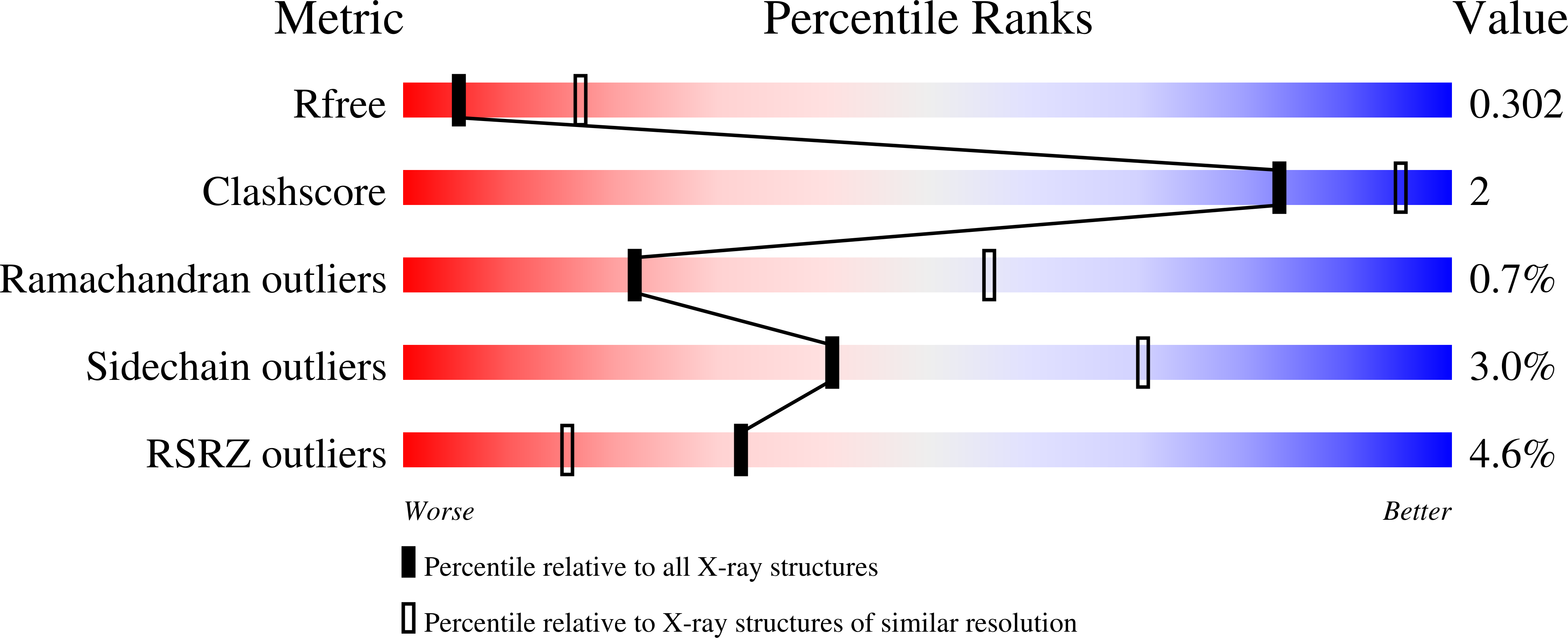
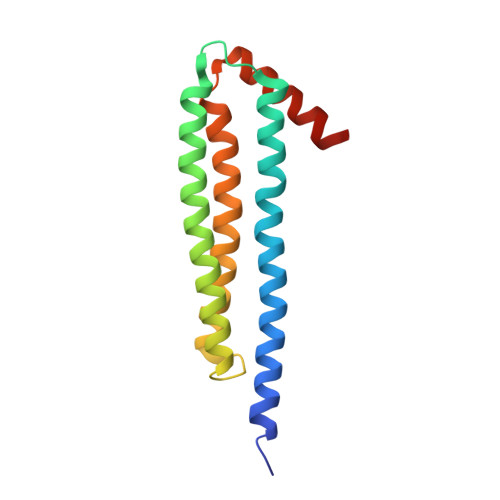
The x-ray structure of NccX, a type II transmembrane metal sensor, from Cupriavidus metallidurans 31A has been determined at a resolution of 3.12 Å. This was achieved after solubilization by dodecylphosphocholine and purification in the presence of the detergent. NccX crystal structure did not match the model based on the extensively characterized periplasmic domain of its closest homologue CnrX. Instead, the periplasmic domains of NccX appeared collapsed against the hydrophobic transmembrane segments, leading to an aberrant topology incompatible with membrane insertion. This was explained by a detergent-induced redistribution of the hydrophobic interactions among the transmembrane helices and a pair of hydrophobic patches keeping the periplasmic domains together in the native dimer. Molecular dynamics simulations performed with the full-length protein or with the transmembrane segments were used along with in vivo homodimerization assays (TOXCAT) to evaluate the determinants of the interactions between NccX protomers. Taken as a whole, computational and experimental results are in agreement with the structural model of CnrX where a cradle-shaped periplasmic metal sensor domain is anchored into the inner membrane by two N-terminal helices. In addition, they show that the main determinant of NccX dimerization is the periplasmic soluble domain and that the interaction between transmembrane segments is highly dynamic. The present work introduces a new crystal structure for a transmembrane protein and, in line with previous studies, substantiates the use of complementary theoretical and in vivo investigations to rationalize a three-dimensional structure obtained in non-native conditions.
Organizational Affiliation:
From the Institut de Biologie Structurale, UMR 5075 CNRS-Commissariat à l'Energie Atomique (CEA)-Université Grenoble-Alpes, 71 Avenue des Martyrs, 38044 Grenoble Cedex 9, France.