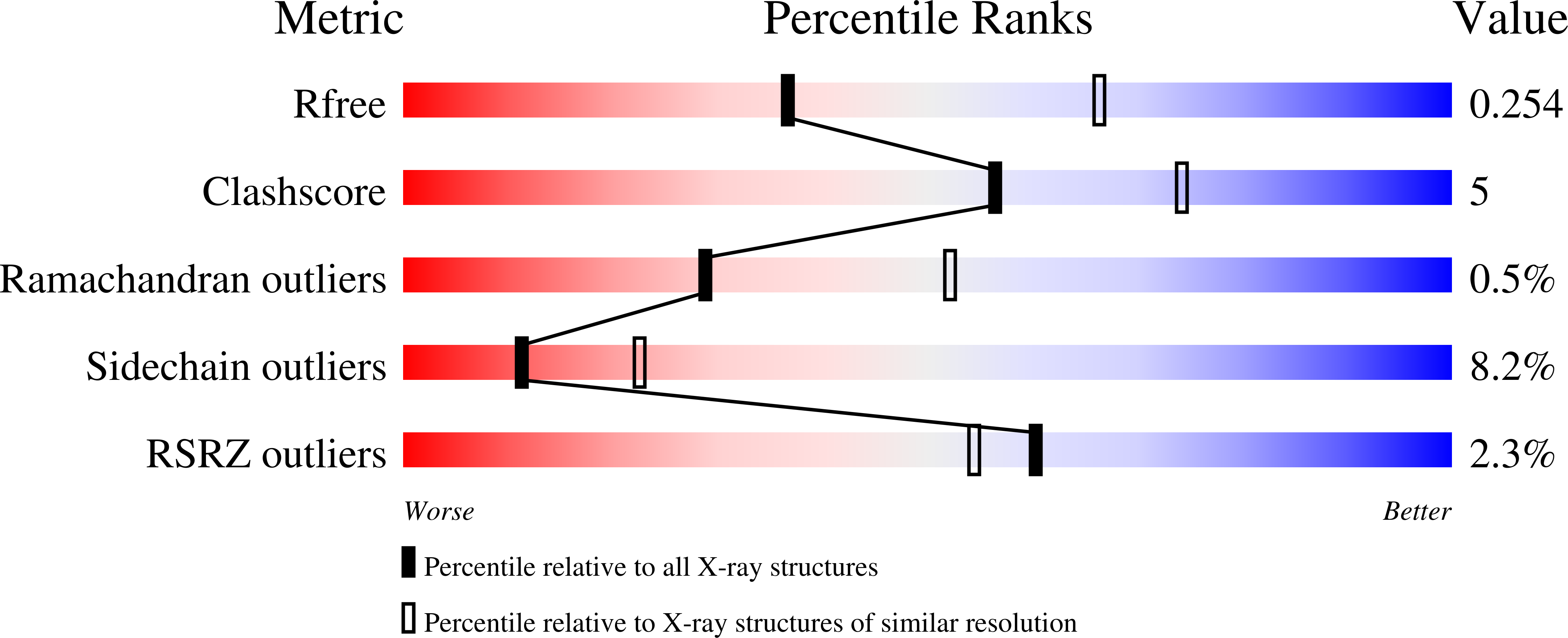
Structural Details of Human Tuba Recruitment by Inlc of Listeria Monocytogenes Elucidate Bacterial Cell-Cell Spreading.
Polle, L., Rigano, L.A., Julian, R., Ireton, K., Schubert, W.(2014) Structure 22: 304
- PubMed: 24332715
- DOI: https://doi.org/10.1016/j.str.2013.10.017
- Primary Citation of Related Structures:
4CC2, 4CC3, 4CC4, 4CC7 - PubMed Abstract:
The human pathogen Listeria monocytogenes is able to directly spread to neighboring cells of host tissues, a process recently linked to the virulence factor InlC. InlC targets the sixth SH3 domain (SH3-6) of human Tuba, disrupting its physiological interaction with the cytoskeletal protein N-WASP. The resulting loss of cortical actin tension may slacken the junctional membrane, allowing protrusion formation by motile Listeria. Complexes of Tuba SH3-6 with physiological partners N-WASP and Mena reveal equivalent binding modes but distinct affinities. The interaction surface of the infection complex InlC/Tuba SH3-6 is centered on phenylalanine 146 of InlC stacking upon asparagine 1569 of Tuba. Replacing Phe146 by alanine largely abrogates molecular affinity and in vivo mimics deletion of inlC. Collectively, our findings indicate that InlC hijacks Tuba through its LRR domain, blocking the peptide binding groove to prevent recruitment of its physiological partners.
Organizational Affiliation:
Department of Biotechnology, University of the Western Cape, Bellville 7535, Cape Town, South Africa; Division of Structural Biology, Helmholtz-Centre for Infection Research, Braunschweig 38124, Germany.