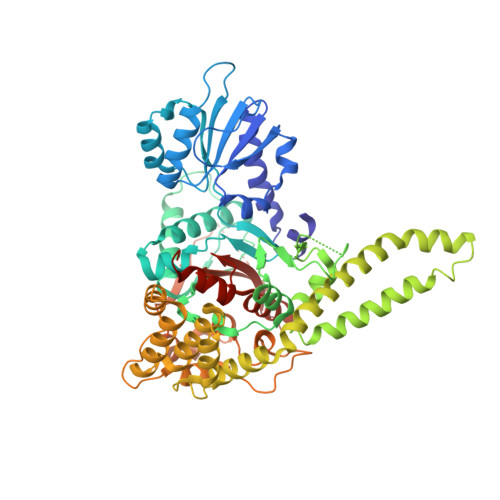
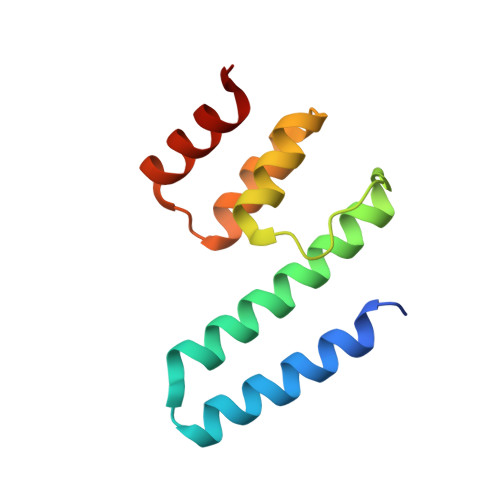
Structural basis of Vps33A recruitment to the human HOPS complex by Vps16.
Graham, S.C., Wartosch, L., Gray, S.R., Scourfield, E.J., Deane, J.E., Luzio, J.P., Owen, D.J.(2013) Proc Natl Acad Sci U S A 110: 13345-13350
- PubMed: 23901104
- DOI: https://doi.org/10.1073/pnas.1307074110
- Primary Citation of Related Structures:
4BX8, 4BX9 - PubMed Abstract:
The multisubunit homotypic fusion and vacuole protein sorting (HOPS) membrane-tethering complex is required for late endosome-lysosome and autophagosome-lysosome fusion in mammals. We have determined the crystal structure of the human HOPS subunit Vps33A, confirming its identity as a Sec1/Munc18 family member. We show that HOPS subunit Vps16 recruits Vps33A to the human HOPS complex and that residues 642-736 are necessary and sufficient for this interaction, and we present the crystal structure of Vps33A in complex with Vps16(642-736). Mutations at the binding interface disrupt the Vps33A-Vps16 interaction both in vitro and in cells, preventing recruitment of Vps33A to the HOPS complex. The Vps33A-Vps16 complex provides a structural framework for studying the association between Sec1/Munc18 proteins and tethering complexes.
Organizational Affiliation:
Department of Clinical Biochemistry, Cambridge Institute for Medical Research, University of Cambridge, Cambridge CB2 0XY, United Kingdom. scg34@cam.ac.uk