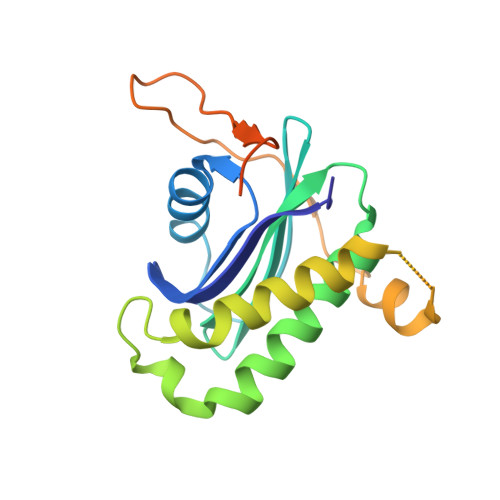
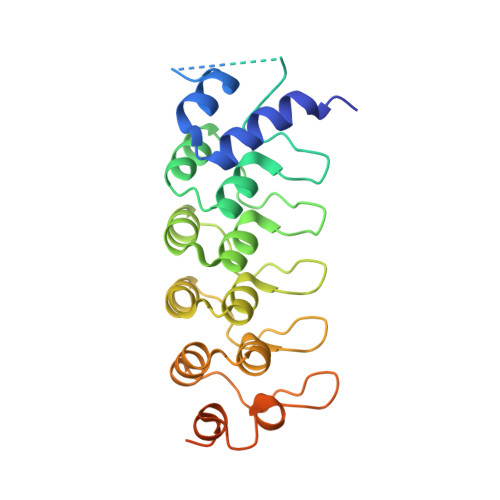
The Binding of Varp to Vamp7 Traps Vamp7 in a Closed, Fusogenically Inactive Conformation.
Schafer, I.B., Hesketh, G.G., Bright, N.A., Gray, S.R., Pryor, P.R., Evans, P.R., Luzio, J.P., Owen, D.J.(2012) Nat Struct Mol Biol 19: 1300
- PubMed: 23104059
- DOI: https://doi.org/10.1038/nsmb.2414
- Primary Citation of Related Structures:
4B93 - PubMed Abstract:
SNAREs provide energy and specificity to membrane fusion events. Fusogenic trans-SNARE complexes are assembled from glutamine-contributing SNAREs (Q-SNAREs) embedded in one membrane and an arginine-contributing SNARE (R-SNARE) embedded in the other. Regulation of membrane fusion events is crucial for intracellular trafficking. We identify the endosomal protein Varp as an R-SNARE-binding regulator of SNARE complex formation. Varp colocalizes with and binds to VAMP7, an R-SNARE that is involved in both endocytic and secretory pathways. We present the structure of the second ankyrin repeat domain of mammalian Varp in complex with the cytosolic portion of VAMP7. The VAMP7-SNARE motif is trapped between Varp and the VAMP7 longin domain, and hence Varp kinetically inhibits the ability of VAMP7 to form SNARE complexes. This inhibition will be increased when Varp can also bind to other proteins present on the same membrane as VAMP7, such as Rab32-GTP.
Organizational Affiliation:
Medical Research Council Laboratory of Molecular Biology, Cambridge, UK.